Background: Plant complex I contains γ-CA subunits whose role is unclear.
Results: 15N labeling and import show CI assembly via a rapidly turning over γ-CA subcomplex.
Conclusion: γ-CAs form an ancient pathway for CI assembly.
Significance: The assembly pathway of complex I in plants is different from that in animals and more closely represents the ancestral enzyme.
Keywords: Bioenergetics/Electron Transfer Complex, Gel Electrophoresis, Mitochondria, Plant Biochemistry, Proteomics, Complex I, γ-Carbonic Anhydrase, Protein Import
Abstract
Subcomplexes of mitochondrial respiratory complex I (CI; EC 1.6.5.3) are shown to turn over in vivo, and we propose a role in an ancestral assembly pathway. By progressively labeling Arabidopsis cell cultures with 15N and isolating mitochondria, we have identified CI subcomplexes through differences in 15N incorporation into their protein subunits. The 200-kDa subcomplex, containing the ancestral γ-carbonic anhydrase (γ-CA), γ-carbonic anhydrase-like, and 20.9-kDa subunits, had a significantly higher turnover rate than intact CI or CI+CIII2. In vitro import of precursors for these CI subunits demonstrated rapid generation of subcomplexes and revealed that their specific abundance varied when different ancestral subunits were imported. Time course studies of precursor import showed the further assembly of these subcomplexes into CI and CI+CIII2, indicating that the subcomplexes are productive intermediates of assembly. The strong transient incorporation of new subunits into the 200-kDa subcomplex in a γ-CA mutant is consistent with this subcomplex being a key initiator of CI assembly in plants. This evidence alongside the pattern of coincident occurrence of genes encoding these particular proteins broadly in eukaryotes, except for opisthokonts, provides a framework for the evolutionary conservation of these accessory subunits and evidence of their function in ancestral CI assembly.
Introduction
NADH-ubiquinone oxidoreductase (complex I (CI),3 EC 1.6.5.3) is the largest (∼1000 kDa) protein complex of the oxidative phosphorylation assembly and is also the major entry point of electrons from the oxidation of NADH into the respiratory chain. CI contains 14 core subunits that represent the minimal active form and that are conserved from prokaryotic to eukaryotic organisms. In the mitochondria of eukaryotes, CI contains an additional 31–38 accessory subunits, a number of which differ among organisms (1–4). Single-particle electron microscope imaging analysis of isolated CI from Arabidopsis and Polytomella identified a specific matrix-exposed domain formed by γ-carbonic anhydrase (γ-CA) subunits that are attached to the membrane arm of mitochondrial CI by the integral membrane protein CA2 (5). In Arabidopsis, this protein domain contains at least three different γ-CA proteins, CA1 (At1g19580), CA2 (At1g47260), and CA3 (At5g66510), having conserved active site regions, and two less well conserved γ-CA-like proteins, CAL1 (At5g66510) and CAL2 (At3g48680), which contain alterations to the primary structure of typical γ-CAs (6). Links between these subunits and photosynthetic function have been postulated (7), a role in photomorphogenesis in plants has been shown by gene knock-out studies (8), and overexpression leads to male sterility in plants (9), but a CA activity from these proteins has not been experimentally confirmed. Although at first these subunits were thought to be specific to CI from photosynthetic organisms (10, 11), evidence has emerged of γ-CA proteins in mitochondria and in isolated CI from a range of other eukaryotic lineages, including slime molds and amoebae that lack photosynthesis (12). Broader phylogenetic studies have also found homology between other CI accessory subunits once thought to be species-or lineage-specific (4).
Given the divergence of CI composition among eukaryotes, both the assembly pathway and the biological importance of accessory subunits in assembly intermediates could be considerably different among major taxonomic groups of eukaryotes. In CI mutants of Neurospora crassa, analysis of CI assembly using radiolabeling pulse-chase has revealed that the matrix and membrane arms assemble independently via separate pathways (13–17). The membrane subunit 20.9, which is absent from mammalian CI, was identified some years ago as a factor essential for assembly of the membrane arm in fungi (18). Based on a combination of radiolabeling pulse-chase experiments, in vitro mitochondrial import and monitoring of tagged CI subunits in assembly-disturbed systems, several CI assembly models have been proposed for human cells (14, 19, 20). In this process, mammalian assembly factors not conserved in other species have been identified (13, 14, 19). Subcomplexes containing CA2 have been reported using antibodies (11) in detergent disassembly studies of CI in plants (21), in Arabidopsis CI subunit knock-out mutants (22), and in wild type Arabidopsis mitochondrial proteomes in saturation mapping of two-dimensional blue native (BN) gels (23). Knockouts of CA2 and CA3 do not yield a clear phenotype in Arabidopsis (5, 11), which might be a consequence of functional redundancy in activities between these proteins. However, suspension cell culture lines derived from the mutant ca2 showed reduced growth rates and lower CI abundance (11), which indicates that γ-CA subunits could be important for CI assembly or structural stability in plants.
However, reports of studies to date have not been able to distinguish between productive and non-productive intermediates, an essential distinction in building a rigorous CI assembly model. Specifically, we needed to determine if subcomplexes were present in vivo or if they were formed in vitro due to detergent solubilization or electrophoresis; if they were incorporated into the small/early assembly intermediates; and if they were productive intermediates that go on to form CI or were simply non-productive subcomplexes that accumulated in mitochondria. We have combined a progressive 15N labeling strategy in vivo (24) with BN polyacrylamide gel electrophoresis (BN-PAGE) and in vitro [35S]Met import and assembly assays to address these issues. This approach has allowed analysis of the incorporation rate of subunits into subcomplexes, mature CI, and supercomplexes of plant mitochondria to investigate their role in assembly and to provide key evidence for the existence of a productive CI subcomplex in vivo in plants. The subunits of the identified assembly subcomplex, lost in animal lineages, are widely conserved in other eukaryotic lineages, providing evidence for their role as ancestral accessory subunits.
EXPERIMENTAL PROCEDURES
Arabidopsis Cell Culture Growth and 15N Labeling
Arabidopsis cell suspension was cultured in growth medium (1× Murashige and Skoog medium without vitamin, 3% sucrose (w/v), 0.5 mg/liter naphthalene acetic acid, 0.05 mg/liter kinetin, pH 5.8) at 22 °C under continuous light conditions with a light intensity of 90 μmol m−2 s−1 and with orbital shaking at 120 rpm. Cultures were maintained in 250-ml Erlenmeyer flasks by the inoculation of 20–25 ml of 7-day-old cells into 100 ml of new medium. The same growth medium without nitrogen (i.e. no ammonium nitrate or potassium nitrate) was used to wash the 7-day-old cell culture three times before it was transferred to 15N growth medium. Ammonium-15N nitrate-15N (1.69 g/liter) and potassium nitrate-15N (1.92 g/liter) (98% 15N; Sigma) were added to growth medium without nitrogen to make the 15N growth medium. Cells labeled for 24, 120, and 168 h with 15N were collected onto filter paper using vacuum filtration. Three biological replicates for each time point were performed.
Protoplast Preparation and Mitochondrial Isolation from Cell Culture
Fresh cells (20–25 g) for each single time point replicate were dispersed in digestion solution (0.4% (w/v) Onozuka cellulase RS, 0.05% (w/v) Kyowa pectolyase Y-23, 0.4 m mannitol, 0.7 g/liter MES, pH 5.7) and gently shaken in the dark for 3 h at room temperature. The solution was centrifuged at 800 × g for 10 min. The supernatant was decanted, and the protoplast pellet resuspended in digestion solution without enzymes by swirling and repeated rounds of centrifugation. Washed protoplasts were then resuspended in extraction buffer (0.4 m sucrose, 50 mm Tris, 3 mm EDTA, and 0.1% BSA (w/v), pH 7.5) and homogenized 5–9 times with a Dounce 0.12-mm homogenizer. Extraction buffer containing broken protoplasts was then centrifuged at 2500 × g for 5 min, the supernatant was recovered, and then this supernatant was subjected to centrifugation at 20,000 × g for 20 min. The resultant pellet was resuspended in 0.5–1 ml of mannitol wash buffer (0.3 m mannitol, 10 mm TES, and 0.1% (w/v) BSA, pH 7.5) using a paint brush, and the suspension was pipetted onto two 35-ml Percoll step gradients comprising 40, 25, and 18% (w/v) Percoll in mannitol wash buffer. The gradients were centrifuged at 40,000 × g for 60 min and decelerated with slow braking. Mitochondria, which form a yellow band at the 40 and 25% interface, were collected and diluted in sucrose wash buffer (0.3 m sucrose, 10 mm TES, pH 7.5). Diluted mitochondria were centrifuged at 24,000 × g for 15 min and decelerated with slow braking. Pellet was collected and diluted again in wash buffer. Washed mitochondria were diluted in 0.5–1 ml of sucrose wash buffer. Isolated mitochondria were quantified with the Bradford assay and were then used for import assays or made into 1-mg protein aliquots and run on BN gels or stored at −80 °C for further experiments.
Arabidopsis Plant Growth and Mitochondrial Preparation
Wild type Arabidopsis (Col-0) or ca2 mutant seeds (∼30 μg) (11) were washed in 70% (v/v) ethanol for 2 min, and then the seeds were placed in a sterilization solution (5% (v/v) bleach, 0.1% (v/v) Tween 20) for 15 min with periodical shaking. Seeds were then washed in sterile water 5 times before being dispensed into a 250-ml plastic vessel containing 80 ml of growth medium (half-strength Murashige and Skoog medium without vitamins, half-strength Gamborg B5 vitamin solution, 5 mm MES, 2.5% (w/v) sucrose, pH 5.8). Arabidopsis seedlings were grown under a 16/8-h light/dark period with light intensity of 100–125 μmol m−2 s−1 at 22 °C for 2 weeks. Ten vessels of wild type or ca2 mutant seedlings were collected for mitochondrial preparations. Seedlings (50–100 g) were homogenized with mortar and pestle, and mitochondria were isolated as described previously (25). The protein content of freshly isolated mitochondria was quantified with a Bradford assay protocol before being used for import experiments.
Two-dimensional BN/SDS-PAGE
Mitochondrial aliquots were washed with 2 ml of sucrose wash buffer (0.3 m sucrose, 10 mm TES, pH 7.5) and then centrifuged at 14,300 × g for 10 min. Pellets were dissolved in 100 μl of digitonin solubilization buffer (30 mm HEPES, pH 7.4, 150 mm potassium acetate, 10% (v/v) glycerol, 50 mg/ml digitonin). Samples were centrifuged at 18,000 × g for 30 min after incubation for 20 min on ice to remove insoluble material and were subsequently supplemented with 5 μl of Serva Blue G250 (5% (w/v) Coomassie Blue in 750 mm aminocaproic acid). Coomassie Blue-treated protein samples were directly loaded onto BN gels. Two-dimensional BN/SDS-PAGE was carried out as described previously (26). Gradient gels (4.5–16% (w/v) acrylamide) were used for the first BN dimension. Second dimension Tricine-SDS-polyacrylamide gels, composed of a 4% stacking gel and a 12% separation gel, were used for separation of protein complex subunits. Electrophoresis parameters and Coomassie staining of BN and BN/SDS-PAGE were carried out as described previously (27).
In-gel Digestion and MALDI-TOF/TOF Analysis
Protein spots in CI+CIII2, CI, and CI subcomplex lanes from BN gels were excised and subjected to in-gel digestion with trypsin using previously described methods (28) but using only a quarter of the trypsin concentration in order to decrease the effect of trypsin autolysis peaks on MS data interpretation. Peptides were analyzed with an UltraFlex III MALDI-TOF/TOF mass spectrometer (Bruker Daltonics). In-gel digested peptides were reconstituted with 5 μl of a solution containing 5% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid (TFA) in double-distilled H2O. Two μl of each sample was spotted onto a MTP 384 MALDI target plate and mixed with 2 μl of spotting matrix (90% acetonitrile, 10% saturated α-cyano-4-hydroxycinnamic acid in TA90 (90% acetonitrile, 0.01% TFA)). Dried spots were overlaid with 10 μl of cold washing buffer (10 mm NH4H2PO4, 0.1% TFA) and allowed to stand for 10 s before removal by pipette. Spots were analyzed at 50–85% laser intensity with up to 1200 shots for MS analysis per spot. Ions between 700 and 4000 m/z were selected for MS/MS experiments using 3% additional laser power. Masses corresponding to trypsin autolysis products were excluded from analysis. A maximum of 15 molecular ions were analyzed by MS/MS from each gel spot. Tandem mass spectrometry data were analyzed using Biotools (Bruker Daltonics) and an in-house Arabidopsis database comprising ATH1.pep (release 9) from the Arabidopsis Information Resource and the Arabidopsis mitochondrial and plastid protein sets (33,621 sequences; 13,487,170 residues), using the Mascot search engine version 2.3.02 and utilizing error tolerances of ±1.2 Da for MS and ±0.6 Da for MS/MS; “Max Missed Cleavages” set to 1; variable modifications of oxidation (Met) and carbamidomethyl (Cys). Only protein matches with more than two peptides and with ion scores greater than 40 were used for analysis (p < 0.05). For all spots, the 0 and 24 h time points were used for both MS and MS/MS analyses for protein IDs, whereas only the MS data were analyzed for 120 and 168 h. Search results were exported to CSV format for each spot.
Quantification of Protein Turnover as NA/(NA + H) and Protein Degradation as KD and Protein Half-life
In order to calculate heavy label incorporation and the relative abundance of NA (natural abundance) and H (heavy labeled) peptide populations, MALDI-TOF/TOF MS data were exported to mzXML files (CompassXport version 3.0 (Bruker)) and were subsequently converted to text files using the ProteomeCommons.org IO Framework version 6.21. Text files were then parsed using a script written in Mathematica version 7.0 (Wolfram Research) for further analysis. Isotopic envelopes were fitted using a two-population model consisting of natural abundance and 15N-labeled fractions with the open source program Isodist (29) as we have described previously (24).
Protein degradation rates (KD) were calculated as described previously (24). The total protein abundance in the cell culture at different time points was quantified using ImageJ of stained protein gels and by Amido Black analysis as described previously (24). An average KD among subunits within CI+III2, CI, and 650- and 200-kDa subcomplexes was separately determined at 24, 120, and 168 h. Half-lives of CI+III2, CI, and 650- and 200-kDa subcomplexes at 24, 120, and 168 h were determined from KD as half-life = ln2/KD. The final KD values and half-lives for CI+III2, CI, and 650- and 200-kDa subcomplexes were determined by averaging the values obtained at 24, 120, and 168 h.
Import in Cell Culture and Plant Mitochondria
The full cDNAs of Arabidopsis thaliana CA1 (At1g19580), CA2 (At1g472660), CA3 (At5g66510), 20.9-(At4g16450), CAL1 (At5g63510), CAL2 (At3g48680), and ATPE (At1g51650) were cloned into pDest14 (Invitrogen) using gateway cloning techniques (Invitrogen) for in vitro transcription and translation as described previously (30). We used imports of the ϵ subunit of ATP synthase (At1g51650), which does not readily get incorporated into native complexes in plant mitochondria, as a control for radiolabeled patterns in the isolated mitochondria. Equivalent quantities of mitochondria isolated from cell culture, wild type, and ca2 knock-out Arabidopsis were used in import reactions as described previously (30).
RESULTS
Higher Turnover Rates of Specific Subunits in the 200- and 650-kDa CI Subcomplexes
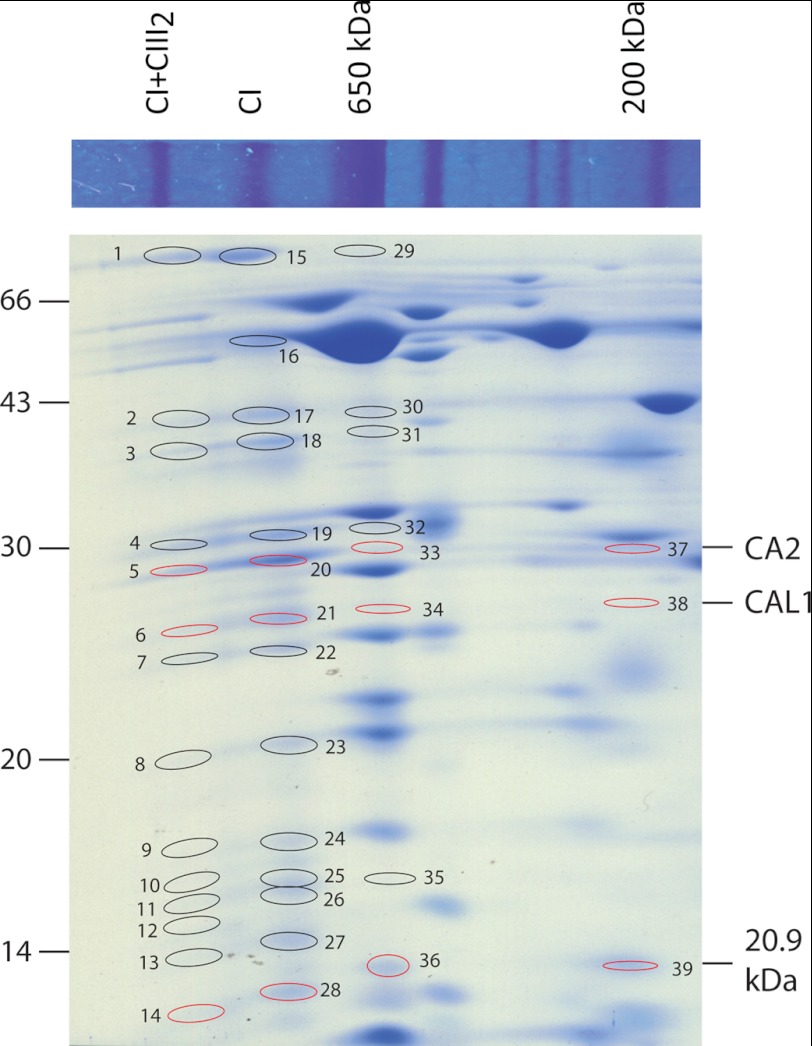
Arabidopsis cells were grown for 24, 120, and 168 h in 98% 15N-enriched MS medium (24), mitochondrial oxidative phosphorylation complexes were prepared and separated by BN-PAGE, and the major protein spots were identified by mass spectrometry. These data were consistent with the reported protein complex proteome of plant mitochondria (23). This confirmed several reported CI subunit-containing protein complexes, including CI+CIII2 and the mature CI holoenzyme (10, 21) (Fig. 1). Peptide mass spectrometry analysis revealed 22 different CI-specific subunits in the CI (ND1, ND7, ND9, 75 kDa, 51 kDa, 39 kDa, 20.9 kDa, 18 kDa, ASHI, B14, B13, B14.7, B16.6, B17.2, CA2, CA3, CAL1, NDU10, NDU12, PDSW, PSST, and TYKY) and 23 different subunits in the CI+CIII2 (ND1, ND7, ND9, 75 kDa, 51 kDa, 39 kDa, 20.9 kDa, 18 kDa, ASHI, B14, B13, B14.7, B16.6, B17.2, CA2, CA3, CAL1, NDU10, NDU11, NDU12, PDSW, PSST, and TYKY). All of these subunits are confirmed subunits in the plant CI holoenzyme (3, 10, 23, 31). Interestingly, the CI subunit CA2 was also found in spot 37, which was a visible stained region in our BN gels (Fig. 1). CI subunits have previously been observed at 650 kDa (22), at 400–450 kDa (3, 22, 23), and at 200 kDa (3, 22, 23) in Arabidopsis mitochondrial BN gel analyses. Analysis of gel regions at these native molecular masses and at masses of the complex I subunits observed in the CI holoenzyme and CI+CIII2 supercomplex was then carried out. This revealed that CI subunits CAL1 and 20.9 kDa could be found along with CA2 at 650 and 200 kDa (Fig. 1 and supplemental Data Set S1). A wider range of protein bands was analyzed, but only those including significant identifications of CI subunits are annotated in Fig. 1.
FIGURE 1.
Coomassie staining of BN and BN/SDS-PAGE separations of mitochondrial membrane protein subunits and identification of complex I subunits. Positions of subunits from CI and CI+CIII2 and the corresponding 650- and 200-kDa subcomplexes are shown. Ellipses and numbers, identified CI subunits (evidence provided in supplemental Data Set S1); red indicates the position of the three noted CI subunits across complexes.
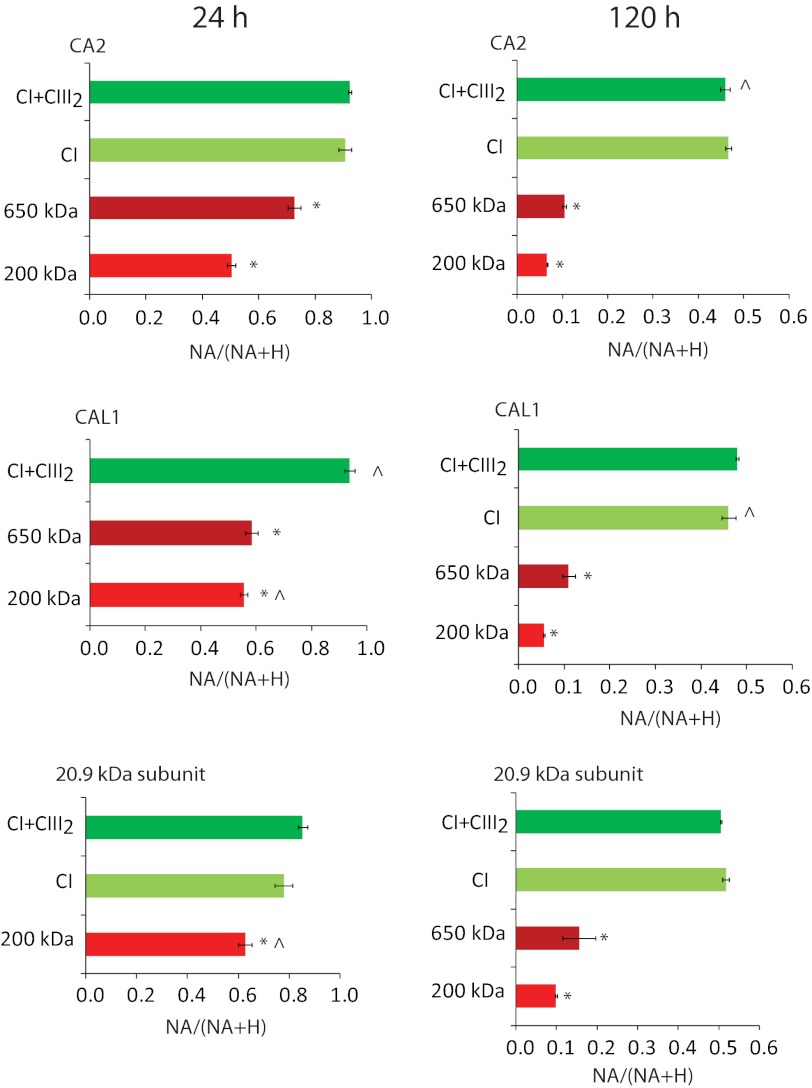
High scoring peptides by the Mascot algorithm (p < 0.05) were further analyzed to determine what proportion of each peptide in mass spectra was derived from the NA of nitrogen in the original cell culture and what proportion derived from the incorporation of 15N (H) into new protein synthesized after the growth medium switch. These values (as outlined under “Experimental Procedures” and in Ref. 24) were used to calculate the NA/(NA + H) ratio for specific peptides from each gel spot. Calculations of NA/(NA + H) for all of the peptides of a given protein were averaged to determine a final NA/(NA + H) ratio ± S.E. This value was used as a proxy for each protein's turnover rate (Fig. 2 and supplemental Data Set S2). Subunits, including CA2, CAL1, and 20.9 kDa, had a significantly lower NA/(NA + H) ratio when the peptides were derived from the 200- and 650-kDa subcomplexes than when the same peptides for the same proteins were derived from CI and CI+CIII2. These results indicate a significantly higher turnover rate for the 200- and 650-kDa subcomplexes (p < 0.05) than for CI and CI+CIII2. These differences were clearly apparent after just 24 h of 15N labeling and were very evident after 120 h of labeling (Fig. 2). Notably, the differences between CI and CI+CIII2 were not statistically significant for any of the three subunits examined (Fig. 2 and supplemental Data Set S2).
FIGURE 2.
Isotope incorporation ratios of subunits in CI-containing complexes after 24 and 120 h of 15N labeling. Shown is the 15N incorporation ratio (NA/(NA + H)) in peptides of CA2, CAL1, and 20.9-kDa subunit at 24 and 120 h. Error bars, S.E. values of biological replicates (n = 3 or else marked with a caret). Different colors show the 200-kDa subcomplex (red), 650-kDa subcomplex (dark red), CI (yellow-green), and CI+CIII2 (green). *, statistically significant lower NA/(NA + H) ratio in the subcomplexes compared with intact CI (24 h CAL1 is compared with CI+CIII2) (p < 0.05, t test). For calculations, see supplemental Data Set S2.
The turnover rates indicated by NA/(NA + H) values do not define the specific contribution of synthesis of 15N protein and the degradation of pre-existing 14N proteins to turnover. However, data on the growth rate of the cell culture allowed us to subsequently calculate the degradation rate (KD) of CI and subcomplexes and convert this to protein half-life. The averaged KD value for the 200-kDa subcomplex (0.42 ± 0.04 day−1) was about 10 times faster than for CI+III2 (0.04 ± 0.01 day−1) and CI (0.04 ± 0.02 day−1) and about 1.6 times faster than for the 650-kDa subcomplex (0.26 ± 0.07 day−1). The averaged protein half-life for the 200-kDa subcomplex (1.7 ± 0.3 day) was thus about 8% of that of CI+III2 (22.0 ± 5.7 days) and CI (23.9 ± 11.9 days) and 60% of that of the 650-kDa subcomplex (3.0 ± 0.7 days). The lower NA/(NA + H) values of subunits in subcomplexes are thus due to their significant in vivo turnover during the course of the 7 days of the experiment rather than just a process of dilution through growth.
Matrix Arm Subunits Have Higher Turnover Rates than Membrane-associated and Mitochondrion-encoded CI Subunits
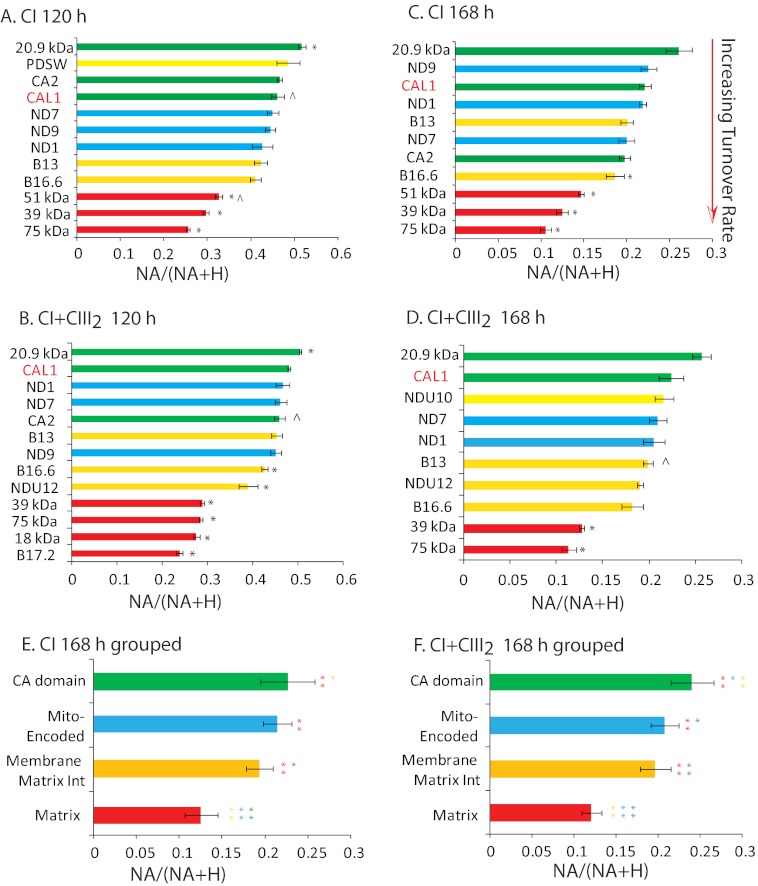
To investigate whether subunits have different turnover rates compared with each other within CI and CI+CIII2, the NA/(NA + H) ratios for a variety of CI subunits after 120 and 168 h of 15N labeling were compared (Fig. 3, A–D). Matrix arm subunits, including the 75-kDa, 51-kDa, 39-kDa, 18-kDa, and B17.2 subunits, exhibited relatively low NA/(NA + H) ratios and thus relatively higher turnover rates than other subunits in both CI and CI+CIII2. The specific matrix-exposed arm components, including CA2, CAL1, and 20.9 kDa, and mitochondrion-encoded subunits, including ND1, ND7, and ND9, had relatively high NA/(NA + H) ratios and thus lower turnover rates. The plant 20.9-kDa subunit, which has been found in both matrix and membrane modules of CI (3, 22), does not have a direct homolog in mammals but is related to fungal NUXM subunits and the 13- and 21-kDa CI subunits in other eukaryotes and was the most stable protein subunit studied in both CI and CI+CIII2.
FIGURE 3.
Isotope incorporation ratios for CI subunits within mature CI and CI+CIII2 after 120 and 168 h of 15N incubation. A–D, the NA/(NA + H) ratio for each subunit is shown for peptides isolated from CI and CI+CIII2 following 120 and 168 h of incubation with 15N. Error bars, S.E. values among biological replicates (n = 3 or else marked with a caret). *, statistical significance (p < 0.05) compared with CAL1. CI subunits are divided into four groups based on colors. Red, matrix group (18 kDa, B17.2, 39 kDa, 51 kDa, and 75 kDa); yellow, membrane and matrix interaction group (B16.6, B13, PDSW, NDU10, and NDU12); blue, mitochondrion-encoded group (ND1, ND7, and ND9); green, γ-CA domain group (CA1, CA2, CAL1, and 20.9 kDa). Error bars, S.E. values among biological replicates (n = 3, or else marked with a caret). *, statistical significance (Student's t test, p < 0.05) compared with the CAL1 subunit. E and F, statistical comparison of the four groups in CI and CI+CIII2 after 168 h of 15N incubation. Error bars, S.D. among measurements of different subunits within a group (n = 6–11). One-way analysis of variance and Tukey's post hoc multiple comparisons among four groups were used; **, p < 0.01; *, p < 0.05. For calculations, see supplemental Data Set S3.
To determine if these observations of varying 15N incorporation rates for different modules of the complex were statistically significant, CI subunits were divided into four groups as detailed in the legend to Fig. 3. One-way analysis of variance and Tukey's post hoc multiple comparisons among the four groups revealed that the matrix group had a significantly higher 15N incorporation rate than every other group (p < 0.01). The membrane/matrix interaction group also had a significantly higher 15N incorporation rate than the specific γ-CA domain group (p = 0.04 in CI, p < 0.01 in I+III2) (Fig. 3, E and F, and supplemental Data Set S3). Notably, there was no difference between the incorporation rate of 15N into the CI holoenzyme and the CI+CIII2 supercomplex, indicating that the two appear to be in equilibrium.
γ-CAs, γ-CALs, and 20.9-kDa Subunit Precursor Proteins Are Imported and Assembled into a Variety of Subcomplexes in Mitochondria
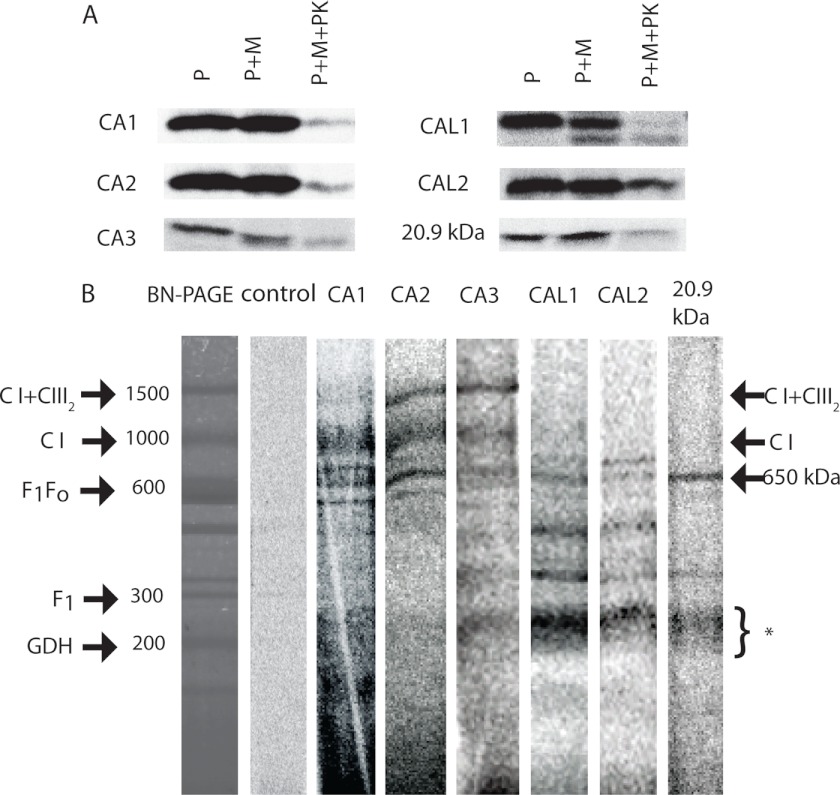
To determine whether γ-CA domain subcomplexes are produced rapidly after import, we performed in vitro import assays with [35S]Met-labeled CA1, CA2, CA3, CAL1, CAL2, and 20.9-kDa subunit precursors into isolated Arabidopsis cell culture mitochondria. Analysis of autoradiographs of SDS-PAGE-separated samples showed that all subunits were imported into mitochondria during a 60-min incubation period and that imported products were resistant to proteinase K (PK) digestion (Fig. 4A). The CAL1 subunit was clearly processed upon import, leading to a ∼2-kDa decrease in its apparent molecular mass, and only the processed forms were PK-protected. The other subunits assayed were not clearly processed, leading to changes in molecular weight, but membrane-bound proteins were PK-protected after 60 min of the import reaction (Fig. 4A). Separation of PK-protected proteins by BN-PAGE revealed that CA3, CAL1 and -2, and 20.9-kDa subunit could be clearly assembled into 200–300-kDa subcomplexes and the 650-kDa subcomplex. CA1–CA3 were assembled to differing degrees into CI and CI+CIII2 (Fig. 4B). This efficient assembly of labeled subunits might explain why the smaller subcomplexes are less defined in CA1 and CA2. Comparison of the radiolabeled import gels with Coomassie-stained gels showed that the 200–300-kDa complexes (Fig. 4B, *) seen in the import experiments were not exactly the same size, when compared with the abundant 200-kDa subcomplex comprising CA2, CAL1, and 20.9-kDa subunits that had been observed to accumulate in Fig. 1; however, they were similar in size to the multiple subcomplexes containing γ-CAs found by CI disassembly (3). The γ-CAL and 20.9-kDa proteins differed with respect to the subcomplexes into which they progressed, compared with the γ-CA proteins. Overall, these results demonstrated that the CA domain subunits were not simply added as accessory components to fully or nearly fully assembled CI, but to different degrees they were present in the full range of subcomplexes that are known to include components of the membrane arm of CI (3, 22). Import of the ϵ subunit of ATP synthase, which fails to assemble into a protein complex, was used as a radiolabeled control import for comparison of labeling on gel images.
FIGURE 4.
Import of CA domain subunits and 20.9-kDa subunit into wild type Arabidopsis mitochondria. A, [35S]Met-labeled precursors (P) were incubated with mitochondria (M) for 60 min with and without PK treatment and were separated by SDS-PAGE. B, [35S]Met-labeled precursors were incubated with fresh mitochondria, following which proteins from PK-treated mitochondria were separated by BN-PAGE. Radiolabeled bands aligned to a Coomassie-stained BN-polyacrylamide gel of the same material and the positions of complexes and subcomplexes are shown. Molecular masses of complexes are reported against standardized masses (23). *, location of 200–300-kDa complex I subcomplexes. The control lane is an equivalent import assay with the ϵ subunit of ATP synthase (At1g51650), which does not readily get incorporated into complexes and serves as a reference for radiolabeling patterns on SDS-polyacrylamide gel images.
CA2 Is Imported and Assembled into a Transient 200-kDa Subcomplex in ca2 Mutant Arabidopsis Mitochondria
To further understand the nature of the γ-CA domain subcomplexes in imports, time series imports were compared between mitochondria isolated from wild type and ca2 knock-out plants that have been previously reported by others (11) and have been propagated in our laboratory. Analysis in the mutant had the potential advantage that [35S]Met-labeled precursors did not have to compete with unlabeled ones, and initial assembly processes and assembly factors might be more abundant in ca2 knockouts, where the abundance of fully assembled CI is reduced (11). A time series of 5, 10, 20, 30, and 60 min of CA2 precursor import was conducted, and the samples were separated by SDS-PAGE and BN-PAGE. More CA2 precursor was imported and PK-protected in mitochondria isolated from Arabidopsis ca2 knockouts than in the equivalent wild type mitochondria (Fig. 5A). On BN gels, it was clear that CA2 precursors were rapidly assembled into a discrete 200-kDa subcomplex in mitochondria from the ca2 knockout (Fig. 5B). In comparison, mitochondria from wild type showed assembly of CA2 into the same 200–300-kDa range of complexes observed during imports into cell culture mitochondria (Fig. 4B versus Fig. 5B). Whereas the [35S]Met incorporation into CI steadily increased in the ca2 knockout during the import time course, the abundance of the 200 kDa band peaked at 20 min and declined thereafter, indicating that it was further assembled into intact CI and was thus a productive intermediate of assembly (Fig. 5B). CI+CIII2 could not be visualized until 20 min, and then it exhibited the same steadily increasing pattern of [35S]Met incorporation seen for CI. A similar but weaker pattern was observed in wild type mitochondria for progressive labeling from subcomplexes to CI and CI+CIII2 (Fig. 5B).
FIGURE 5.
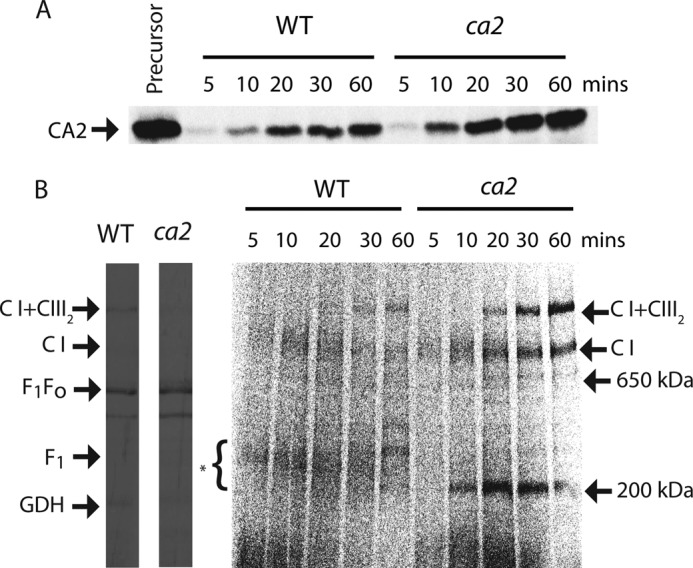
Import of CA2 into ca2 mutant mitochondria. Mitochondria were prepared from 2-week-old hydroponically grown wild type and ca2 knock-out Arabidopsis. A, CA2 precursors were incubated with wild type and ca2 knock-out mitochondria for 5, 10, 20, 30, and 60 min; mitochondria were then treated with PK; and mitochondrial proteins were separated by SDS-PAGE. B, mitochondria were treated with PK, and solubilized components were then separated by BN-PAGE. Radiolabeled bands aligned to Coomassie-stained BN-polyacrylamide gels of the same samples, and the positions of complexes and subcomplexes are indicated. *, location of 200–300-kDa complex I subcomplexes.
Conservation and Losses of 20.9, γ-CA, and γ-CAL Proteins among Eukaryotic Lineages
Although γ-CAs were initially considered plant-specific subunits of CI (6, 10, 11), more recent work has demonstrated the widespread occurrence of γ-CA homologs throughout eukaryotes, with the notable exception of opisthokonts (animals + fungi) (12). Extending this analysis, we have found that in organisms in which deep genome and/or transcriptome sequencing has been performed, multiple γ-CA genes are often found (Table 1 and supplemental Data Set S4). Where the inferred protein sequences are deemed to be complete at the N terminus, many of these γ-CAs have a predicted mitochondrial localization (Table 1). A bioinformatic survey to assess the phylogenetic distribution of the 20.9-kDa protein sequence shows a similarly wide occurrence of the corresponding gene across the various eukaryotic supergroups (Table 1). However, in contrast to what is seen with animals, the 20.9-kDa subunit is very widely present in fungi (where it has a known role in CI assembly (18). Outside of opisthokonts, γ-CAs and the 20.9-kDa subunit exhibit a strikingly similar pattern of co-occurrence (Table 1). Aside from Arabidopsis, direct evidence for the presence of both γ-CAs and the 20.9-kDa subunit in mitochondrial CI is available for the green alga, Chlamydomonas reinhardtii (32), as well as Acanthamoeba castellanii (33). Various of these components have also been identified in proteomic screens of purified mitochondria from additional non-photosynthetic protists, including Tetrahymena thermophila (34), Dictyostelium discoideum (35), and Trypanosoma brucei (36, 37) (Table 1). Evidently, the subunits of the subcomplex described here in Arabidopsis are widely conserved and coincident in CI across eukaryotes.
TABLE 1.
Phylogenetic distribution of γ-CA and 20.9-kDa subunits of CI
Major taxonomic groups are based on Fig. 2 of Brown et al. (51). Shaded and boldface entries indicate that there is direct evidence for one or more of the corresponding components in mitochondria and/or isolated CI, as described under “Results” (further details are also provided in supplemental Data Set S4). +/−, presence/absence of the 20.9-kDa subunit gene in a given organism. Data in the table are taken largely from publicly available genome/transcriptome sequencing projects, where gene coverage is likely to be high. Additional sequences for other organisms as well as other information (accession numbers, mitochondrial targeting probabilities) may be found in supplemental Data Set S4.
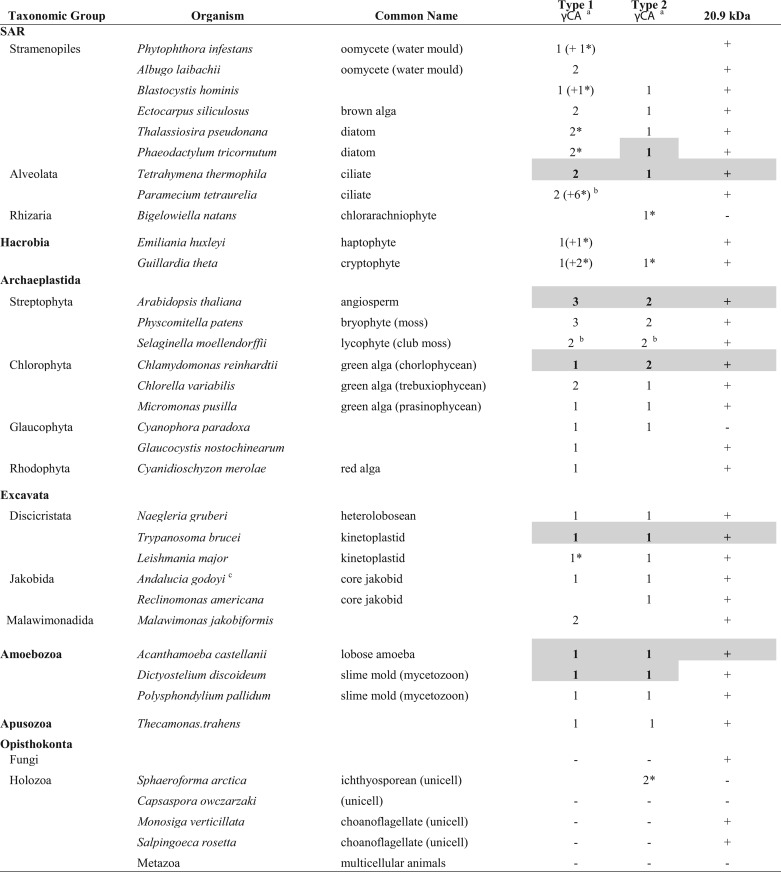
a γ-CA homologs can be classified into two types, based on a high degree of conservation (Type 1) or not (Type 2) in the N-terminal region (12). Plant γ-CAs fall into the Type 1 category, whereas plant γ-CALs are included among Type 2 γ-CAs. An asterisk denotes that the γ-CA Type assignment is tentative.
b γ-CA multiplicity attributable to genome duplication(s).
c Unpublished data from the A. godoyi genome project.
DISCUSSION
CI Matrix Arm and Mitochondrion-encoded Subunits Have Different Turnover Rates
The assembly of the largely nucleus-encoded matrix arms and the set of mitochondrion-encoded proteins that are mainly in the membrane arm of CI occur via independent processes in other organisms (13–17, 19, 20, 38); thus, it is conceivable that these subcomplexes might turn over at different rates in plants. Our data show that the matrix arm (including members of both the N-module (51 kDa and 75 kDa) and the Q-module (39 kDa), as defined for the mammalian complex (39)) have a higher turnover rate than either the mitochondrion-encoded subunits or the γ-CA domain components of the membrane arm. Elevated protein turnover can arise from high synthesis or high degradation rates or a combination of both processes (24). As matrix arm subunits form the major ROS production sites in the respiratory chain, these components may be damaged more readily and require replacement more frequently. ROS-mediated protein oxidation can sensitize proteins to proteolytic attack (40, 41). It is possible that the ROS released from CI could lead to increased oxidation of subunits in the vicinity or that ROS damage in the matrix is generally greater in magnitude than in the membrane (41). The matrix arm subunits are also likely to be more accessible to matrix proteases than the mainly membrane subunits. It is also possible that the natural breakdown of intact CI simply begins with the degradation of matrix arm subunits, irrespective of damage or accessibility. Whichever the reason, the higher turnover rate of matrix arm subunits than of both the mitochondrion-encoded and the specific γ-CA domain subunits demonstrates that different functional modules of CI have distinct protein half-lives in plant cells.
Supercomplex Formation Does Not Significantly Stabilize Arabidopsis Complex I
It has previously been proposed that formation of supercomplex I+III2 might stabilize complex I in Arabidopsis (42). Lowering protein turnover by supercomplex formation could provide a functional significance to these superassemblies in the electron transport chain, beyond their role in electron transport catalysis. This proposal is supported by evidence from mice that knockout of complex III (43) or complex IV (44) subunits decreases the content of complex I, suggesting that supercomplex formation stabilizes complex I. It has also been proposed in human mitochondria that supercomplex formation is required for assembly of complex I as well as for its stabilization (45). In addition, structural stabilization of labile membrane protein complexes is proposed as a major function of supercomplex formation in Paracoccus denitrificans (46). As we pointed out under “Results,” however, we did not show any impact of supercomplex formation on protein turnover rate in Arabidopsis cells (Fig. 3). Hence, these direct measurements are not able to confirm a role for supercomplex formation in enhanced stabilization of complex I. Rather, it appears that plant complex I is in equilibrium between the complex I and the supercomplex I+III2 states. Higher order complexes and respirasomes could not be detected in our analysis of Arabidopsis membranes, so we are not able to determine if these might have enhanced stability, but this technique could be used in systems where larger supercomplexes are evident and provide in vivo evidence of relative stability.
A Specific Subcomplex of Unknown Function Proteins Has a Productive Role in CI Assembly
A range of knock-out mutants for CI subunits leading to the accumulation of certain intermediates has previously been exploited to propose that CA2 is incorporated at the early stage of CI assembly (22), and γ-CA-containing subcomplexes have been detected in BN-polyacrylamide gels of wild type mitochondria (23). Using a 15N labeling approach, we have shown the higher 15N incorporation rate of subunits contained in subcomplexes than in CI and CI+CIII2, revealing that these structures were not created in vitro by solubilization or electrophoresis artifacts but rather are in vivo intermediates of disassembly or assembly of CI.
The CI subcomplexes could conceivably be disassembly intermediates that have a high degradation rate. However, the higher 15N incorporation in subcomplexes as compared with CI and CI+CIII2 means this explanation only holds if dissociation of the complex is biased toward newly assembled CI or CI+CIII2. It is not clear what could cause such a bias. Also, our calculations based on Coomassie staining abundance together with the proportion of peptide 15N reveal that the amount of 15N-labeled subunits within subcomplexes exceeds the total amount of 15N label in both CI and CI+CIII2 combined. Hence, the balance of the data, from 15N incorporation alone, is in favor of the 200-kDa subcomplexes found in wild type being assembly rather than disassembly intermediates.
The import experiments using γ-CAs, γ-CALs, and 20.9-kDa subunits showed that all of these subunits can be imported into cell culture mitochondria and that they form a series of subcomplexes ranging in size from 100 to 300 kDa, although the 200-kDa complex found in wild type did not predominate (Fig. 1 versus Figs. 4 and 5). Imported [35S]Met-labeled precursors must compete with the corresponding subunits present in vivo and could lessen the subcomplex signal or even change the original assembly process, especially when redundant subunits compete with subunits having high sequence similarity. Mitochondria from CI subunit mutants have proved to be a useful system for detection of assembly intermediates in mammals (14, 19). We found that more CA2 precursor could be imported into ca2 mutant Arabidopsis mitochondria and that a strong 200-kDa subcomplex band could be detected, which weakened after 20 min as the radiolabel was transferred into CI and CI+CIII2. Thus, γ-CAs and γ-CALs appear to assemble into a productive membrane 200-kDa subcomplex, which serves as the initiator of CI assembly. Our data provide key experimental evidence to justify the model recently proposed in plants (22) that subunits (including at least ND1 and ND7 and matrix subunits of 75 and 39 kDa) are added to a plant-specific 200-kDa subcomplex to form a 650-kDa subcomplex that is further assembled into CI.
Our data do not prove that CA, CAL, and the 20.9-kDa subunit are the only subunits of this 200-kDa subcomplex. Previous surveys of BN-PAGE have found evidence for At2g28430 and ND9 in this region on BN gels (23). Although we were not able to confirm this in our data, the possibility of other CI subunits in this 200-kDa complex cannot be excluded and is likely given its mass. However, the low abundance of the complex has not allowed us to date to make any progress in finding other rapid turnover subunits in this mass range.
Conservation of the Subunits of 200-kDa Subcomplex Suggests an Ancestral Feature of CI Important for CI Assembly
The patterns of conservation of genes for γ-CAs and the 20.9-kDa subunit and the evidence for their conserved presence in CI (Table 1) argue that both protein types are ancestral components of CI, present at the earliest stages of eukaryotic cell evolution; notably, however, this co-occurrence pattern is not preserved in opisthokonts (fungi + animals) (Table 1). Of particular note is the finding that several protists included within the assemblage Holozoa (animals and their specific unicellular relatives (47)) are seen to encode either γ-CAs or the 20.9-kDa subunit, although (like fungi) not both (Table 1). Intriguingly, genes for both γ-CAs and the 20.9-kDa subunit can also be retrieved from the recently determined genome sequence of the apusomonad Thecamonas trehans, arguably the closest neighbor to opisthokonts (48). These observations indicate that progressive loss of components of this putative assembly subcomplex began in opisthokonts at the base of this lineage, after its divergence from its closest related lineages, Apusozoa and Amoebozoa. In Holozoa, loss of all of the subunits known to be in the subcomplex appears to have been complete before the emergence of multicellular animals (Metazoa). Outside of eukaryotes, γ-CAs are prominent among α-proteobacteria, and the γ-CAs of these organisms are the most similar in sequence to the eukaryotic γ-CAs, although in this case they do not appear to be associated with CI (49). Considering the α-proteobacterial ancestry of mitochondria (50), it is possible (perhaps probable) that this bacterial group was the evolutionary source of mitochondrial γ-CAs, which were then recruited to serve in CI assembly early in eukaryotic cell evolution.
Supplementary Material
Acknowledgments
We thank Dr. Ian Castleden (Centre for Computational Systems Biology, University of Western Australia) for expertise and assistance in file format conversions and concatenation. We also thank Dr. Marek Elias (Charles University, Prague) for permission to include unpublished data from the on-going Andalucia godoyi genome sequencing project (Table 1 and supplemental Data Set S4).
This work was supported by Australian Research Council (ARC) Centre of Excellence in Plant Energy Biology Grant CE0561495 (to A. H. M. and J. W.).

This article contains supplemental Data Sets 1–4.
- CI
- complex I
- CA
- carbonic anhydrase
- CAL
- carbonic anhydrase-like
- BN
- blue native
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- TES
- 2-[(2-Hydroxy-1,1-bis(hydroxymethyl)ethyl)amino]ethanesulfonic acid
- ROS
- reactive oxygen species
- NA and H peptide
- natural abundance and heavy labeled peptide, respectively.
REFERENCES
- 1. Walker J. E., Skehel J. M., Buchanan S. K. (1995) Structural analysis of NADH: ubiquinone oxidoreductase from bovine heart mitochondria. Methods Enzymol. 260, 14–34 [DOI] [PubMed] [Google Scholar]
- 2. Videir A., Duarte M. (2001) On complex I and other NADH:ubiquinone reductases of Neurospora crassa mitochondria. J. Bioenerg. Biomembr. 33, 197–203 [DOI] [PubMed] [Google Scholar]
- 3. Klodmann J., Sunderhaus S., Nimtz M., Jänsch L., Braun H. P. (2010) Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22, 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cardol P. (2011) Mitochondrial NADH:ubiquinone oxidoreductase (complex I) in eukaryotes. A highly conserved subunit composition highlighted by mining of protein databases. Biochim. Biophys. Acta 1807, 1390–1397 [DOI] [PubMed] [Google Scholar]
- 5. Sunderhaus S., Dudkina N. V., Jänsch L., Klodmann J., Heinemeyer J., Perales M., Zabaleta E., Boekema E. J., Braun H. P. (2006) Carbonic anhydrase subunits form a matrix-exposed domain attached to the membrane arm of mitochondrial complex I in plants. J. Biol. Chem. 281, 6482–6488 [DOI] [PubMed] [Google Scholar]
- 6. Perales M., Parisi G., Fornasari M. S., Colaneri A., Villarreal F., González-Schain N., Echave J., Gómez-Casati D., Braun H. P., Araya A., Zabaleta E. (2004) γ carbonic anhydrase like complex interact with plant mitochondrial complex I. Plant Mol. Biol. 56, 947–957 [DOI] [PubMed] [Google Scholar]
- 7. Zabaleta E., Martin M. V., Braun H. P. (2012) A basal carbon concentrating mechanism in plants? Plant Sci. 187, 97–104 [DOI] [PubMed] [Google Scholar]
- 8. Wang Q., Fristedt R., Yu X., Chen Z., Liu H., Lee Y., Guo H., Merchant S. S., Lin C. (2012) The γ-carbonic anhydrase subcomplex of mitochondrial complex I is essential for development and important for photomorphogenesis of Arabidopsis. Plant Physiol. 160, 1373–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Villarreal F., Martín V., Colaneri A., González-Schain N., Perales M., Martín M., Lombardo C., Braun H. P., Bartoli C., Zabaleta E. (2009) Ectopic expression of mitochondrial γ carbonic anhydrase 2 causes male sterility by anther indehiscence. Plant Mol. Biol. 70, 471–485 [DOI] [PubMed] [Google Scholar]
- 10. Heazlewood J. L., Howell K. A., Millar A. H. (2003) Mitochondrial complex I from Arabidopsis and rice. Orthologs of mammalian and fungal components coupled with plant-specific subunits. Biochim. Biophys. Acta 1604, 159–169 [DOI] [PubMed] [Google Scholar]
- 11. Perales M., Eubel H., Heinemeyer J., Colaneri A., Zabaleta E., Braun H. P. (2005) Disruption of a nuclear gene encoding a mitochondrial γ carbonic anhydrase reduces complex I and supercomplex I + III2 levels and alters mitochondrial physiology in Arabidopsis. J. Mol. Biol. 350, 263–277 [DOI] [PubMed] [Google Scholar]
- 12. Gawryluk R. M., Gray M. W. (2010) Evidence for an early evolutionary emergence of γ-type carbonic anhydrases as components of mitochondrial respiratory complex I. BMC Evol. Biol. 10, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Küffner R., Rohr A., Schmiede A., Krüll C., Schulte U. (1998) Involvement of two novel chaperones in the assembly of mitochondrial NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 283, 409–417 [DOI] [PubMed] [Google Scholar]
- 14. Mimaki M., Wang X., McKenzie M., Thorburn D. R., Ryan M. T. (2012) Understanding mitochondrial complex I assembly in health and disease. Biochim. Biophys. Acta 1817, 851–862 [DOI] [PubMed] [Google Scholar]
- 15. Schulte U. (2001) Biogenesis of respiratory complex I. J. Bioenerg. Biomembr. 33, 205–212 [DOI] [PubMed] [Google Scholar]
- 16. Tuschen G., Sackmann U., Nehls U., Haiker H., Buse G., Weiss H. (1990) Assembly of NADH:ubiquinone reductase (complex I) in Neurospora mitochondria. Independent pathways of nuclear-encoded and mitochondrially encoded subunits. J. Mol. Biol. 213, 845–857 [DOI] [PubMed] [Google Scholar]
- 17. Videira A., Duarte M. (2002) From NADH to ubiquinone in Neurospora mitochondria. Biochim. Biophys. Acta 1555, 187–191 [DOI] [PubMed] [Google Scholar]
- 18. Schulte U., Fecke W., Krüll C., Nehls U., Schmiede A., Schneider R., Ohnishi T., Weiss H. (1994) In vivo dissection of the mitochondrial respiratory NADH:ubiquinone oxidoreductase (complex I). Biochim. Biophys. Acta 1187, 121–124 [DOI] [PubMed] [Google Scholar]
- 19. Lazarou M., McKenzie M., Ohtake A., Thorburn D. R., Ryan M. T. (2007) Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol. Cell. Biol. 27, 4228–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vogel R. O., van den Brand M. A., Rodenburg R. J., van den Heuvel L. P., Tsuneoka M., Smeitink J. A., Nijtmans L. G. (2007) Investigation of the complex I assembly chaperones B17.2L and NDUFAF1 in a cohort of CI-deficient patients. Mol. Genet. Metab. 91, 176–182 [DOI] [PubMed] [Google Scholar]
- 21. Klodmann J., Lewejohann D., Braun H. P. (2011) Low-SDS Blue native PAGE. Proteomics 11, 1834–1839 [DOI] [PubMed] [Google Scholar]
- 22. Meyer E. H., Solheim C., Tanz S. K., Bonnard G., Millar A. H. (2011) Insights into the composition and assembly of the membrane arm of plant complex I through analysis of subcomplexes in Arabidopsis mutant lines. J. Biol. Chem. 286, 26081–26092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klodmann J., Senkler M., Rode C., Braun H. P. (2011) Defining the protein complex proteome of plant mitochondria. Plant Physiol. 157, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li L., Nelson C. J., Solheim C., Whelan J., Millar A. H. (2012) Determining degradation and synthesis rates of Arabidopsis proteins using the kinetics of progressive 15N labeling of two-dimensional gel-separated protein spots. Mol. Cell Proteomics 11, DOI: 10.1074/mcp.M111.010025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee C. P., Eubel H., O'Toole N., Millar A. H. (2008) Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol. Cell Proteomics 7, 1297–1316 [DOI] [PubMed] [Google Scholar]
- 26. Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 27. Eubel H., Jänsch L., Braun H. P. (2003) New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiol. 133, 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taylor N. L., Heazlewood J. L., Day D. A., Millar A. H. (2005) Differential impact of environmental stresses on the pea mitochondrial proteome. Mol. Cell Proteomics 4, 1122–1133 [DOI] [PubMed] [Google Scholar]
- 29. Sperling E., Bunner A. E., Sykes M. T., Williamson J. R. (2008) Quantitative analysis of isotope distributions in proteomic mass spectrometry using least-squares Fourier transform convolution. Anal. Chem. 80, 4906–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carrie C., Giraud E., Duncan O., Xu L., Wang Y., Huang S., Clifton R., Murcha M., Filipovska A., Rackham O., Vrielink A., Whelan J. (2010) Conserved and novel functions for Arabidopsis thaliana MIA40 in assembly of proteins in mitochondria and peroxisomes. J. Biol. Chem. 285, 36138–36148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meyer E. H., Taylor N. L., Millar A. H. (2008) Resolving and identifying protein components of plant mitochondrial respiratory complexes using three dimensions of gel electrophoresis. J. Proteome Res. 7, 786–794 [DOI] [PubMed] [Google Scholar]
- 32. Cardol P., Vanrobaeys F., Devreese B., Van Beeumen J., Matagne R. F., Remacle C. (2004) Higher plant-like subunit composition of mitochondrial complex I from Chlamydomonas reinhardtii. 31 conserved components among eukaryotes. Biochim. Biophys. Acta 1658, 212–224 [DOI] [PubMed] [Google Scholar]
- 33. Gawryluk R. M., Chisholm K. A., Pinto D. M., Gray M. W. (2012) Composition of the mitochondrial electron transport chain in Acanthamoeba castellanii. Structural and evolutionary insights. Biochim. Biophys. Acta 1817, 2027–2037 [DOI] [PubMed] [Google Scholar]
- 34. Smith D. G., Gawryluk R. M., Spencer D. F., Pearlman R. E., Siu K. W., Gray M. W. (2007) Exploring the mitochondrial proteome of the ciliate protozoon Tetrahymena thermophila. Direct analysis by tandem mass spectrometry. J. Mol. Biol. 374, 837–863 [DOI] [PubMed] [Google Scholar]
- 35. Czarna M., Mathy G., Mac'Cord A., Dobson R., Jarmuszkiewicz W., Sluse-Goffart C. M., Leprince P., De Pauw E., Sluse F. E. (2010) Dynamics of the Dictyostelium discoideum mitochondrial proteome during vegetative growth, starvation and early stages of development. Proteomics 10, 6–22 [DOI] [PubMed] [Google Scholar]
- 36. Acestor N., Ziková A., Dalley R. A., Anupama A., Panigrahi A. K., Stuart K. D. (2011) Trypanosoma brucei mitochondrial respiratome. Composition and organization in procyclic form. Mol. Cell Proteomics 10, DOI: 10.1074/mcp.M110.006908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Panigrahi A. K., Ogata Y., Ziková A., Anupama A., Dalley R. A., Acestor N., Myler P. J., Stuart K. D. (2009) A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9, 434–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ugalde C., Vogel R., Huijbens R., van den Heuvel B., Smeitink J., Nijtmans L. (2004) Human mitochondrial complex I assembles through the combination of evolutionary conserved modules. A framework to interpret complex I deficiencies. Hum. Mol. Genet. 13, 2461–2472 [DOI] [PubMed] [Google Scholar]
- 39. Hunte C., Zickermann V., Brandt U. (2010) Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329, 448–451 [DOI] [PubMed] [Google Scholar]
- 40. Grune T., Reinheckel T., Davies K. J. (1997) Degradation of oxidized proteins in mammalian cells. FASEB J. 11, 526–534 [PubMed] [Google Scholar]
- 41. Bender T., Leidhold C., Ruppert T., Franken S., Voos W. (2010) The role of protein quality control in mitochondrial protein homeostasis under oxidative stress. Proteomics 10, 1426–1443 [DOI] [PubMed] [Google Scholar]
- 42. Dudkina N. V., Eubel H., Keegstra W., Boekema E. J., Braun H. P. (2005) Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc. Natl. Acad. Sci. U.S.A. 102, 3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Acin-Pérez R., Bayona-Bafaluy M. P., Fernández-Silva P., Moreno-Loshuertos R., Pérez-Martos A., Bruno C., Moraes C. T., Enríquez J. A. (2004) Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13, 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Diaz F., Fukui H., Garcia S., Moraes C. T. (2006) Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell. Biol. 26, 4872–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Calvaruso M. A., Willems P., van den Brand M., Valsecchi F., Kruse S., Palmiter R., Smeitink J., Nijtmans L. (2012) Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum. Mol. Genet. 21, 115–120 [DOI] [PubMed] [Google Scholar]
- 46. Stroh A., Anderka O., Pfeiffer K., Yagi T., Finel M., Ludwig B., Schägger H. (2004) Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. J. Biol. Chem. 279, 5000–5007 [DOI] [PubMed] [Google Scholar]
- 47. Lang B. F., O'Kelly C., Nerad T., Gray M. W., Burger G. (2002) The closest unicellular relatives of animals. Curr. Biol. 12, 1773–1778 [DOI] [PubMed] [Google Scholar]
- 48. Cavalier-Smith T., Chao E. E. (2010) Phylogeny and evolution of Apusomonadida (Protozoa: Apusozoa). New genera and species. Protist 161, 549–576 [DOI] [PubMed] [Google Scholar]
- 49. Yip C. Y., Harbour M. E., Jayawardena K., Fearnley I. M., Sazanov L. A. (2011) Evolution of respiratory complex I. “Supernumerary” subunits are present in the α-proteobacterial enzyme. J. Biol. Chem. 286, 5023–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gray M. W., Burger G., Lang B. F. (1999) Mitochondrial evolution. Science 283, 1476–1481 [DOI] [PubMed] [Google Scholar]
- 51. Brown M. W., Kolisko M., Silberman J. D., Roger A. J. (2012) Aggregative multicellularity evolved independently in the eukaryotic supergroup Rhizaria. Curr. Biol. 22, 1123–1127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.