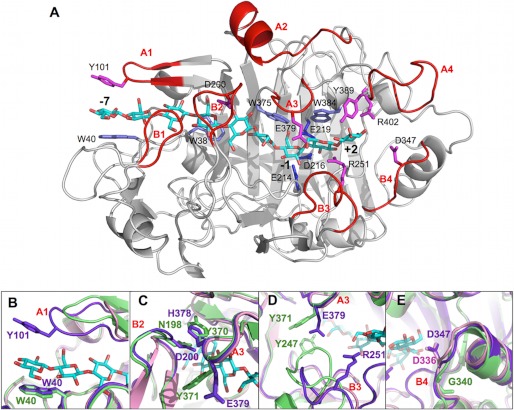
FIGURE 4.
A, overall structure of the HirCel7A enzyme (SX5 structure) with a docked ligand from the H. jecorina 8CEL structure (17). The ligand is shown in cyan, and loops and residues of interest are labeled. B, superposition of loop A1 at the tunnel entrance over subsite −7 from HirCel7A (violet), HjeCel7A (green), and PchCel7D (pink). The HirCel7A loop A1 contains a tyrosine residue (Tyr-101) not present in HjeCel7A or PchCel7D. Loop A1 in PchCel7D is significantly truncated. C, superposition of loop A3 and loop B2 over the −4 subsite. The enzymes exhibit different loop-loop contacts and sequence diversity. D, superposition of loops A3 and B3 near the catalytic center subsite −1 and the product sites +1/+2. HjeCel7A exhibits a longer loop B3, which forms stable contacts to loop A3 across the catalytic center. The HirCel7A loop B3 is two residues shorter and interacts via water with Glu-379 on loop A3. PchCel7D exhibits the most exposed catalytic center because of a six-residue deletion in loop B3. E, superposition of loop B4 shows the aspartate residue that may interact with the reducing end of the product at subsite +2, which is present in HirCel7A (Asp-347, D347) and PchCel7D (Asp-336, D336) but is deleted in HjeCel7A.