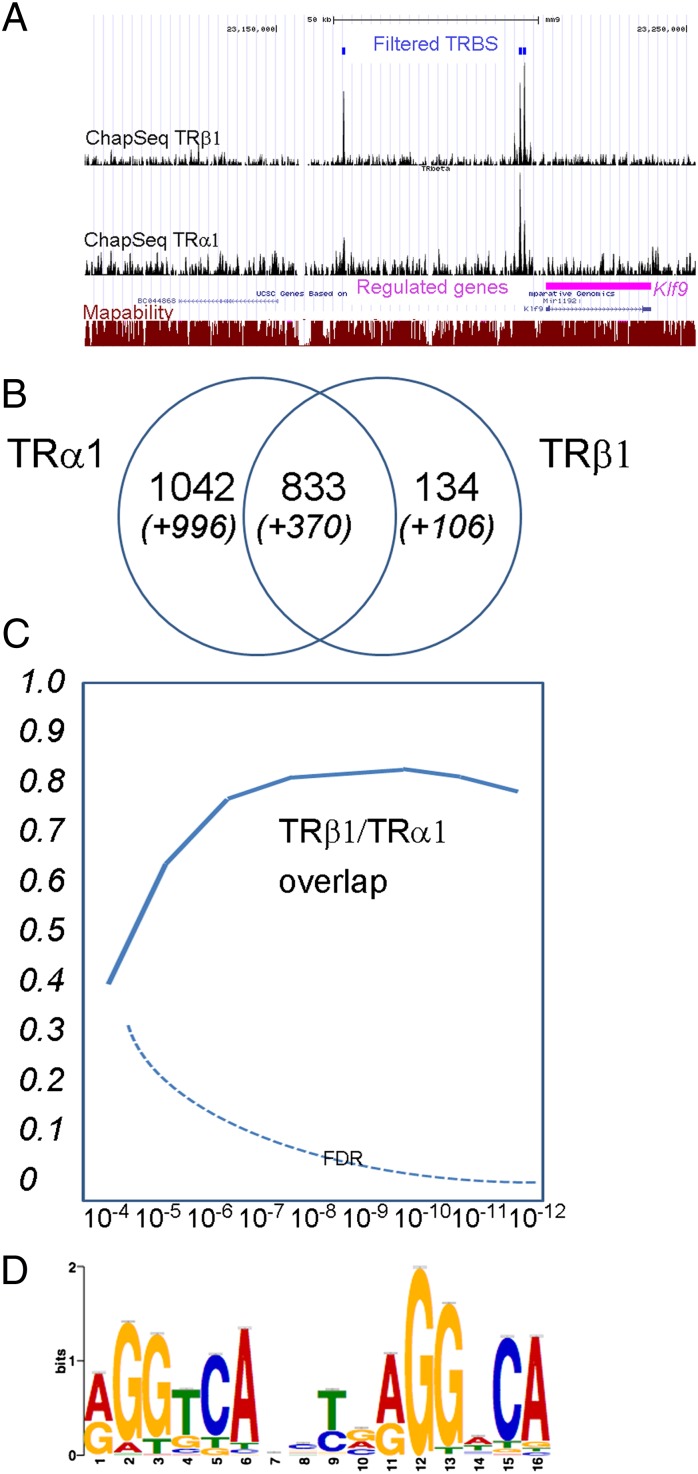
Fig. 5.
TRα1 and TRβ1 cistromes. (A) representative view of the UCSC mouse genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway) around the Klf9 locus. (Bottom) Klf9 position and exon/intron composition are indicated by boxes (exons) and bars (introns). Mapability for 36-mer tags is indicated as a red line ranging from 0 (not mapable) to 1 (fully mapable) for every position. Each ChAP-Seq experiment is represented as a signal track, providing the number of tags sequenced on 10-bp sliding windows (scale on the right-hand side is the number of counted tags). Two shared TRBS and one binding site detected only for TRα1 are located upstream of Klf9. Note that for Klf9, the TRBS are not at the previously reported position (57). (B) Venn diagrams representing the number of binding sites for each ChAP/ChIP-Seq experiment and the number of shared TRBS. The threshold P value is 10−7 for the sites identified in a single experiment. Additional sites (in parentheses and italics) are the ones observed in more than one experiment, using a 10−4 threshold for peak calling, taking into account the data of the RXR ChIP-Seq experiment (Fig. S3). (C) Limited overlap between TRα1 and TRβ1 cistromes. Depending on the P value used for peak calling, the fraction of TRβ1-binding sites overlapping with TRα1-binding site varies. However, unlike what is observed between TRα1 and RXR (Fig. S3), overlap never exceeds 0.8 even for P value <10−7. The false discovery rate was calculated by assuming that all TRα1-binding sites should be also identified in the RXR ChIP-Seq experiment performed on C17.2α cells. (D) Consensus sequence defined by the CHIP-MEME algorithm using all of the TRBS is close to a direct repeat with a 4-nt spacer (DR4). According to structure analysis, RXR recognizes the 5′ half-site (5′AGGTCA) and TR recognizes the 3′ half-site (5′AGGNCA).