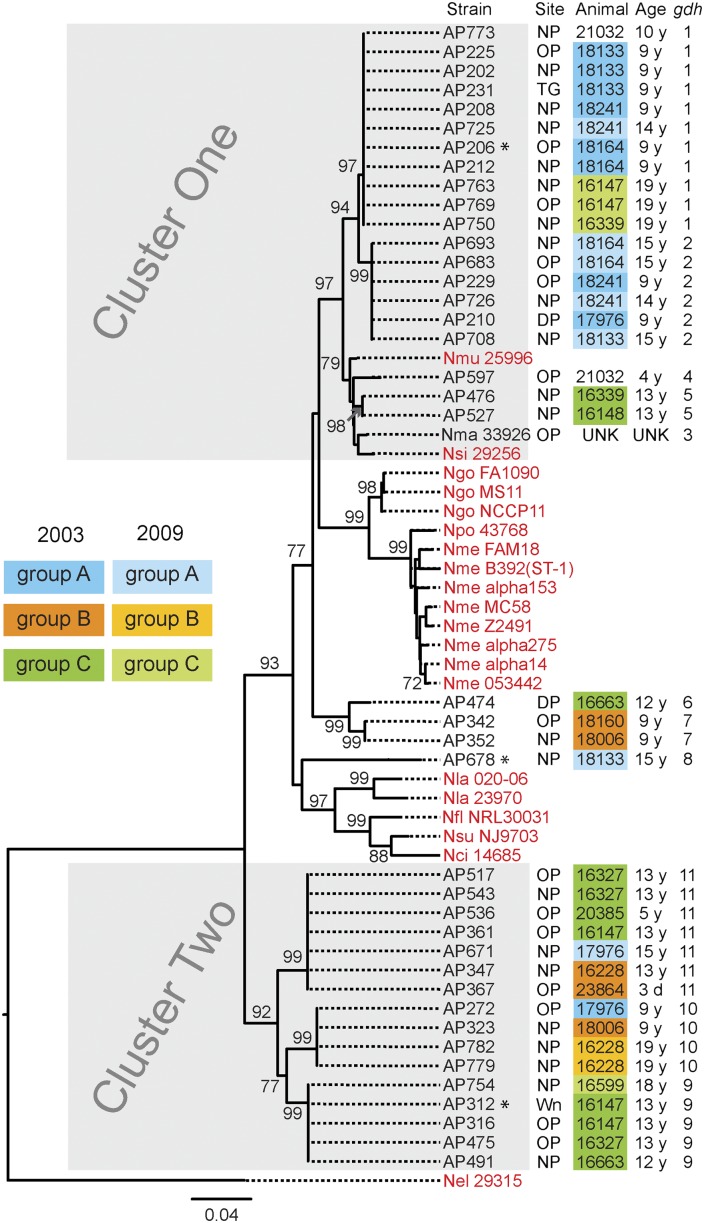
Fig. 1.
gdh sequences resolve RM Neisseria into two clusters. A rooted neighbor-joining tree constructed with gdh sequences from 40 RM Neisseria isolates (present study), N. macacae (Nma; ATCC 33926), and 20 human-adapted Neisseria (in red). Shown for each RM Neisseria strain are its strain number, its site of isolation, the animal from which it was cultured, the age of the animal, and its gdh allele number. Cluster one and cluster two RM Neisseria strains are in gray boxes. Housing groups of the animals are color coded: blue, group A; orange, group B; and green, group C. Neisseria strains cultured from animals in the same housing group is identified by the housing group color. Darker shade of a color indicates the 2003 survey; lighter shade indicates the 2009 survey. Three strains marked with an asterisk were inoculated into animals during this study. DP, dental plaque; Nci, N. cinerea (ATCC 14685); Nel, N. elongata (ATCC 29315); Nfl, N. flavescens (NRL30031); Ngo, N. gonorrohoeae (FA1090, MS11 and NCCP11); Nla, N. lactamica (20-060, ATCC 23970); Nme, N. meningitidis (FAM18, B392, α153, MC58, Z2491, α275, α14, and 053422), Nmu, N. mucosa (Nmu ATCC 25996); NP, nasopharynx; Npo, N. polysaccharea (Npo ATCC 43768); Nsi, N. sicca (ATCC 29256); Nsu, N. subflava (NJ9703); OP, oropharynx; TG, tongue; Wn, wound. Bootstrap values >70% are indicated (2,000 replications).