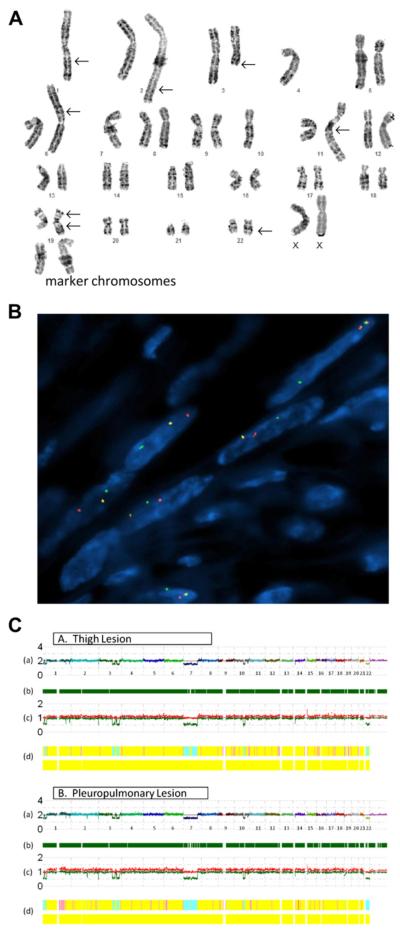
Figure 2.
A) Conventional karyotype of the pleuropulmonary benign metastasizing leiomyoma revealing a hypodiploid clone characterized by loss of chromosomes 1, 4, 7, and 10; gain of 2–5 marker chromosomes; additional material of unknown origin on 1q, 2q, 6p, 11q, 19p, and 22q; a deletion of 3q; and a derivative 19 with additional unknown material on both long and short arms. (B) FISH analysis with a HMGA1 break-apart probe demonstrates split red and green signals indicative of a rearrangement of this locus. (C) SNP array whole genome view of the thigh and pleuropulmonary lesions (A and B, respectively) showing nearly identical genomic profiles, with the exception of an additional loss of 2q22.1 in the pleuropulmonary lesion but not the thigh lesion; (A) estimated copy number as log2 ratio–chromosomes are color coded and sequential along the x-axis; (B) dark green bars represent heterozygous reads; (C) allele-based analysis (high and low alleles are shown in red and green, respectively); and (D) hidden Markov model estimates for copy number (top bar: pink = 3, yellow = 2, aqua = 1) and LOH likelihood (bottom bar: negative for the presence of LOH).