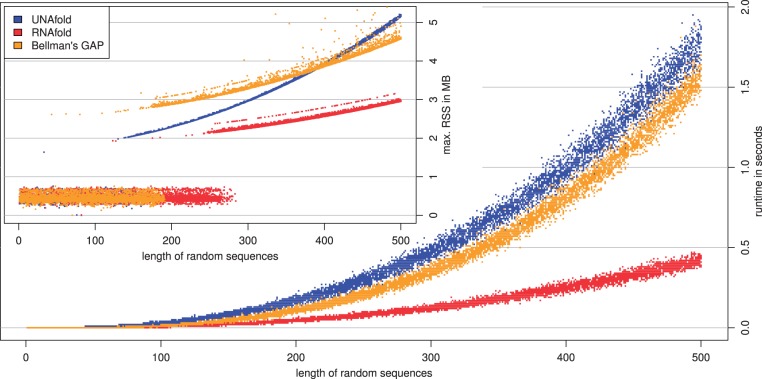
Fig. 2.
Measurement on 10 000 uniformly distributed random sequences with length between 1 and 500 bases. See text for discussion. Runtime (user + system) and memory consumption (max, RSS) measured by UNIX tool ‘memtime’ (by Johan Bengtsson). RNAfold version 2.0.2 -d 1 –noLP –noPS, UNAfold: hybrid-ss-min –suffix = DAT –mfold –NA = RNA –tmin = 37 –tinc = 1 –tmax = 37 –sodium = 1 –magnesium = 0 -I, Bellman’s GAP: mfe * dotBracket –backtrace -t microstate grammar. Short sequence memory consumption seems to fit into initial stack/heap size of an OS process