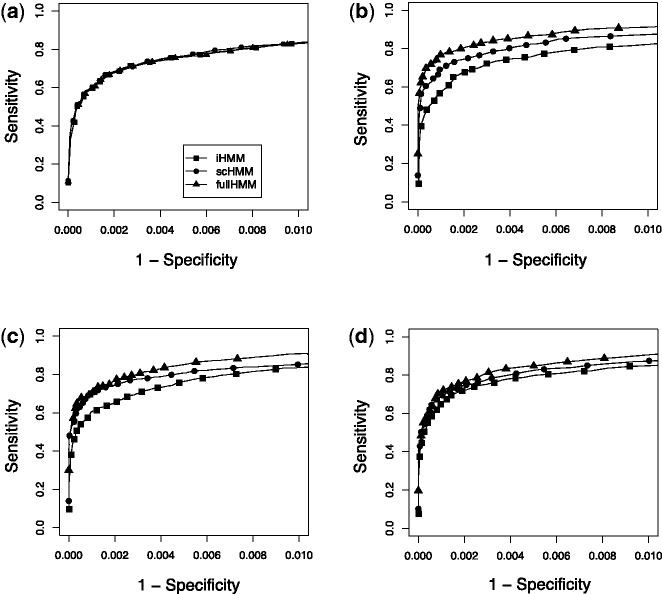
Fig. 2.
Simulation studies. Each method is represented by different symbols: squares for iHMM, circles for scHMM and triangles for fullHMM. (a) Independent case (2-fold): short-length signals were planted in random locations in three different series data. (b) One-group case (2-fold): replicate experiments where binding sites are expected to be shared in all experiment. (c) Two-group case (2-fold): two sets of three correlated series. (d) Three-group case (2-fold): three inter-dependent groups of two correlated series. In all panels, signal was simulated from Poisson (10) and background noise was simulated from Poisson (5)