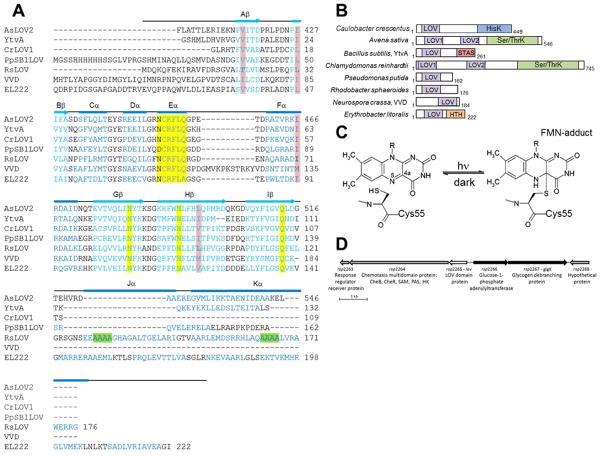
Figure 1.
LOV domain homology and chemistry (A) Structure-based sequence alignment of LOV domains, Avena sativa LOV2 (2V1A), Bacillus subtilis (2PR5), Chlamydomonas reinhardtii (1N9L), Pseudomonas putida (3SW1), Rhodobacter sphaeroides, Neurospora crassa Vivid. (2PD7), and Erythrobacter litoralis (3P7N). Secondary structure elements noted above (α-helical residues in blue, β-sheet in aqua). Residues critical to flavin coordination are highlighted in yellow, sites of RLOV single residue variants are highlighted in pink, and the four Ala repeat in Jα and Kα helices are highlighted in green. (B) Domain arrangement of sequence aligned LOV domain proteins (number of residues specified). HisK, histidine kinase; Ser/ThrK, serine/threonine kinase; STAS, sulfate transporter anti-s antagonist; HTH, helix-turn-helix. (C) Schematic of light-induced cysteinyl-FMN (C4a) covalent adduct formation. (D) Genetic context of LOV domain protein in R. sphaeroides.