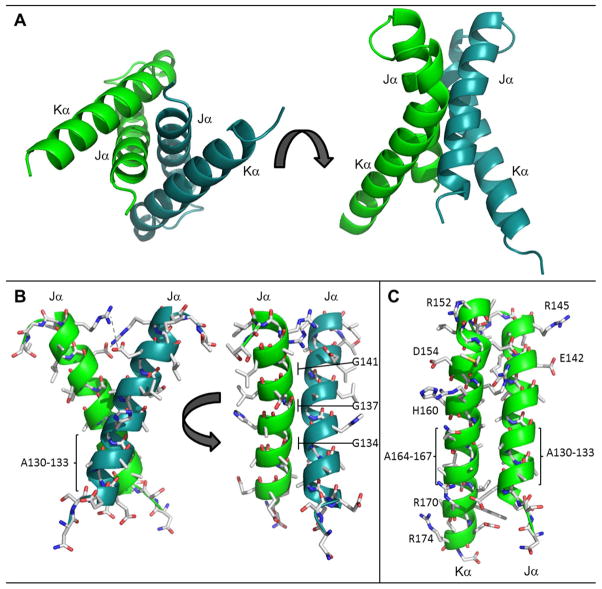
Figure 4.
The RsLOV dimerization motif. (A) Four helix bundle at interface of the RsLOV crystallographic dimer, A and B subunits distinguished by color. (B) Side chain contacts at Jα interface of chain A and chain B (C) Side chain contacts at the Jα and Kα interface. There are salt bridge between Arg145 and Glu142 (same monomer) and between Arg15 and Glu154 (opposing monomer). Hydrogen bonding interactions range from 2.45 to 3.66 Å and also include the amide H of Arg158 to the backbone O of Gly17 (opposing monomer), and the amide H of Arg159 to the side chain O of Ser124 (opposing monomer). The residues with the most buried surface area are within the crossing point of the two Jα helices at the region of the four alanine repeat: Ala130–133, Gly134, Gly137, Ala138, and the cross-section of Kα including Glu154, Arg158, and Ala162.