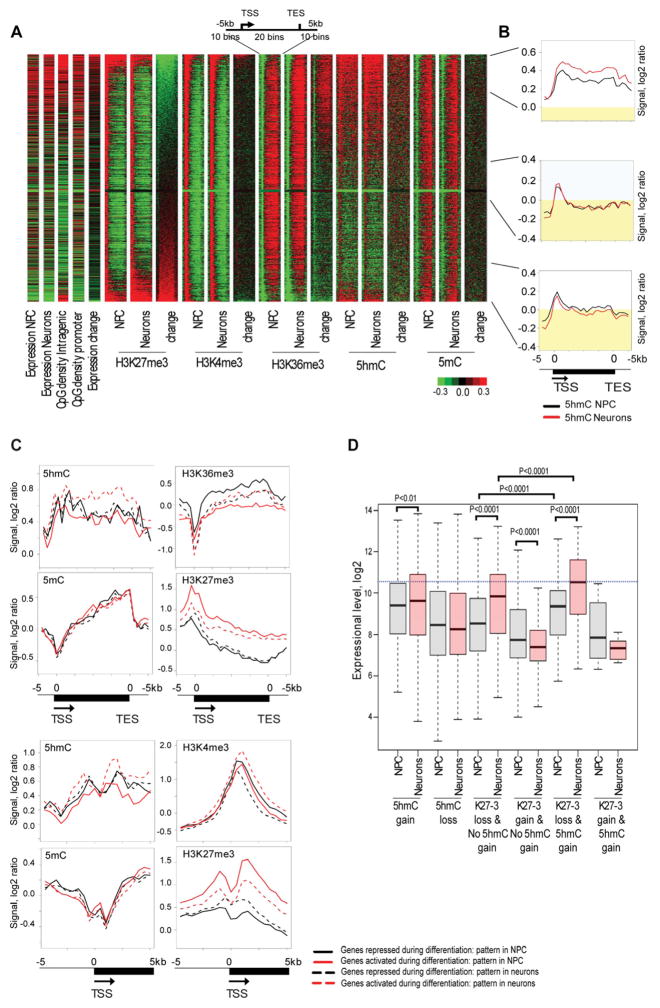
Figure 2. Gain of intragenic 5hmC is associated with loss of H3K27me3 during neuronal differentiation.
A. Heatmap analysis of 5mC, 5hmC and histone methylation marks. The heat-map for chromatin modifications and their changes during neuronal differentiation was generated for genes larger than 2 kb and represents a region containing the gene body and the surrounding area (−5 kb to [gene body] to +5 kb). The heat-map contains information about expression in NPCs, expression in neurons, intragenic CpG density, promoter CpG density, expression changes during neuronal differentiation, histone modifications, 5hmC and 5mC patterns for the analyzed cell type and changes of these epigenetic marks during neuronal differentiation (neurons versus NPC). All analyzed genes were sorted by intragenic H3K27me3 changes. Green color indicates a low level or loss, black represents no change and red specifies an increase or high level of a mark. The top 15% of genes with the greatest loss of intragenic H3K27me3, 15% of genes with an intermediate state of H3K27me3 changes and top 15% of the genes with highest gain of intragenic H3K27me3 during neuronal differentiation are indicated on the right.
B. Composite profiles of the 5hmC mark in the top 15% of genes with greatest loss of intragenic H3K27me3, 15% of genes with intermediate state of H3K27me3 changes and top 15% of the genes with highest gain of intragenic H3K27me3 during neuronal differentiation (from top to bottom). The black line indicates 5hmC levels in undifferentiated cells and red color represents 5hmC in neurons. Yellow indicates 5hmC signal below zero.
C. Epigenetic changes associated with differential expression of genes marked by 5hmC. This analysis was done for genes, which have 5hmC enrichment in the gene body and/or promoters and belong to the top 25% activated or top 25% repressed genes during neuronal differentiation. Composite profiles were generated for promoter regions (−5 kb to TSS to +5 kb) and entire genes (−5 kb to TSS to gene body to TES plus 5 kb). Red solid lines indicate the status of epigenetic marks in NPCs for genes, which are activated during neuronal differentiation. Red dotted lines reflect the status of epigenetic marks in neurons for genes, which are activated during differentiation. Black solid lines indicate the status of epigenetic marks in NPCs for genes, which are repressed during differentiation. Black dotted lines reflect the status of epigentic marks in neurons for genes, which are repressed during differentiation.
D. Gain of intragenic 5hmC and loss of H3K27me3 characterizes genes activated during neuronal differentiation. Gene expression levels in NPCs and in neurons were plotted for six groups of genes: genes which gained intragenic 5hmC during neuronal differentiation, genes which lost intragenic 5hmC, genes which lost H3K27me3 at the TSS and its adjacent intragenic region (−0.5 kb<TSS<4.5 kb) and did not gain 5hmC, genes which gained H3K27me3 at the TSS and its adjacent intragenic region and did not gain intragenic 5hmC, genes which gained intragenic 5hmC and lost H3K27me3 at the TSS and its adjacent intragenic region and genes which gained intragenic 5hmC with simultaneous gain of H3K27me3 at promoters. P-values for significant differences between groups (t-test) are shown.