Figure 4. Intergenic NMIs are associated with distal regulatory elements, non-coding RNAs, and unannotated transcripts.
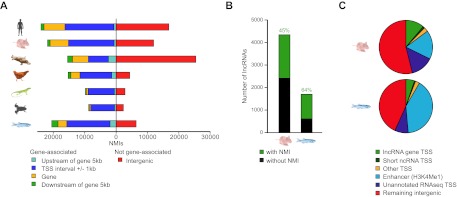
(A) Most NMIs are associated with known protein-coding genes (left) but a substantial proportion are located within intergenic regions of the genome (right). (B) NMIs (green) are found at 45% and 64% of all known long non-coding RNA (lncRNA) TSSs (black) in mouse and zebrafish respectively. (C) A pie chart depicting the proportion of intergenic NMIs (>5 kb from a protein-coding gene) associated with different genomic features in mouse embryonic stem (ES) cells and zebrafish 24 hpf embryos. The association was performed hierarchically in the following order: lncRNA TSSs, other non-coding RNA TSSs (miRNAs, rRNAs, snRNAs, or snoRNAs), other TSSs (pseudogenes and processed transcripts), putative enhancer mark H3K4me1 and novel RNA-seq TSSs. This analysis indicates that intergenic NMIs mark novel transcriptional units or regulatory elements.
