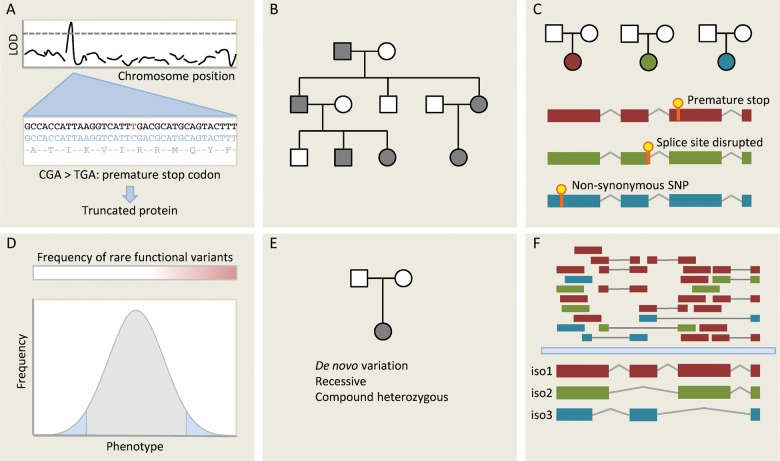
Figure 3.
Different approaches for gene discovery in humans using next-generation sequencing (NGS). (A) One of the simplest applications of NGS is deep sequencing genes at a known disease locus to identify functional variants that may be responsible for the observed effect. Here, a C/T substitution generates a novel stop codon, which truncates a gene product that may be functionally important. (B) Mendelian disease within a single family is typically genetically homogenous: in the absence of phenocopies affected individuals will all share the same causative variant. Whole-genome or whole-exome sequencing can be used to identify functional variants segregating with disease in a family. Many variants will be shared through simple relatedness, so large families are needed for this approach. It also makes assumptions about which classes of variants are likely to underlie Mendelian diseases—typically truncating variants and very rare non-synonymous SNPs. If the causative variant is synonymous or non-coding then it is unlikely to be detected by targeted NGS approaches. (C) An alternative methodology is to sequence unrelated individuals with the same phenotype or endophenotype. Here, we do not expect that affected individuals will carry the same variant, but we hypothesize that they may carry distinct variants in the same gene. This is a powerful approach for genetically homogenous conditions, but inherited cardiac conditions are typically heterogenous (variation in many different genes yields the same phenotype) limiting the applicability of this approach. (D) Strategy C can be extended to continuous traits. If rare variants of moderately large effect contribute to the phenotype then we may be able to detect these by focusing sequencing efforts on the extremes of a very large population, seeking genes that are enriched for rare functional variants using burden testing. (E) Where disease is caused by de novo variation, this may be detected by sequencing trios (proband and both parents). This approach may also be applied to recessive phenotypes. (F) RNA sequencing not only measures total transcript abundance, but can also quantify different isoforms, detect novel transcripts and novel splicing events, and identify sequence variants. Here, isoform 1 is the predominant transcript, but RNAseq provides evidence for two other isoforms. This can be used in integrative genomic studies.