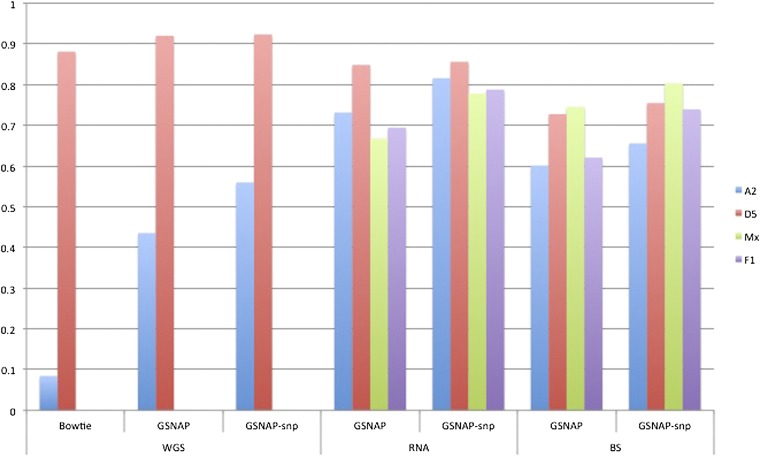
Figure 4 .
Mapping efficiency with and without SNP-tolerant mapping. Reads were mapped by Bowtie (WGS only), GSNAP, and GSNAP with SNP-tolerant mapping (GSNAP-snp). WGS reads from G. arboreum (A2), G. raimondii (D5), were mapped to the reference genome of G. raimondii. Subsequently, RNA-seq and BS-seq reads from A2, D5, G. hirsutum (Mx) and the F1 diploid hybrid (F1) also mapped using SNP-tolerant mapping.