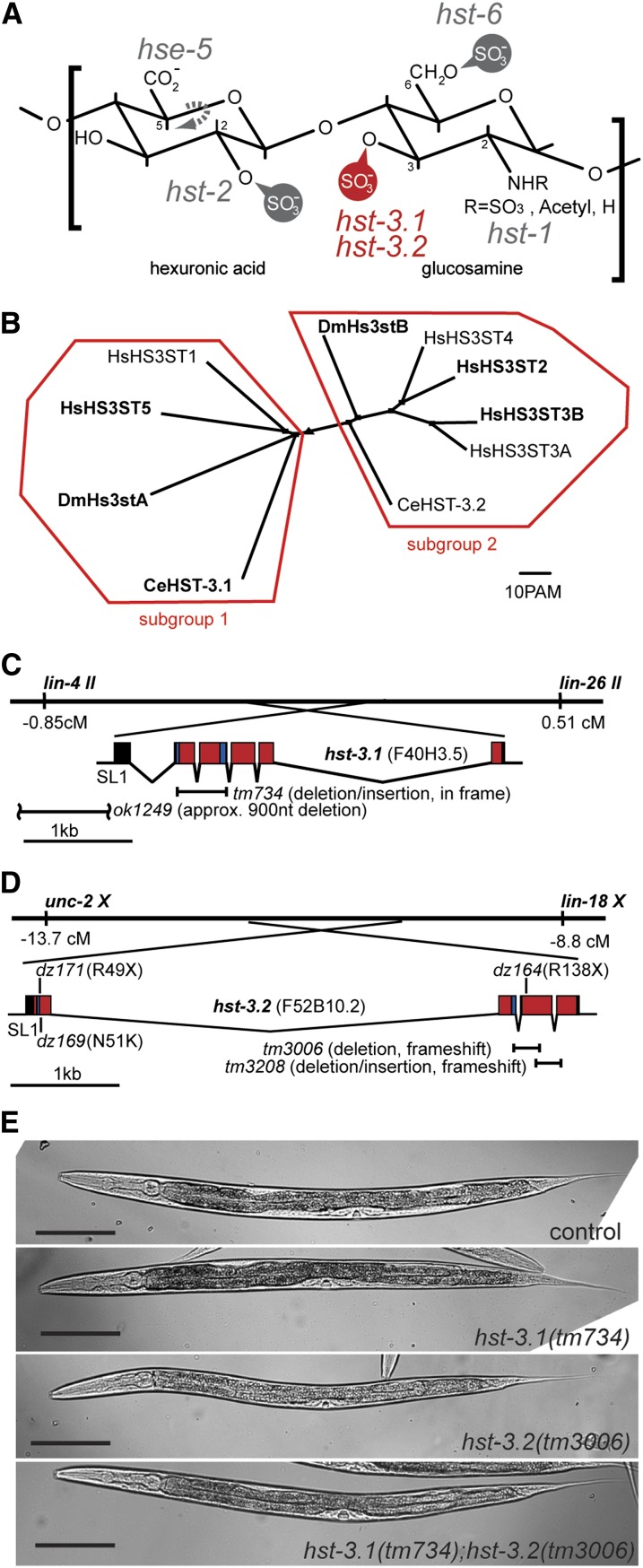
Figure 1 .
HS 3-O-sulfotransferases in C. elegans. (A) Characteristic HS disaccharide comprising a hexuronic acid (glucuronic acid or iduronic acid) and glucosamine. The C. elegans genes coding for HS modification enzymes are indicated next to the positions they modify: hse-5, HS C5 glucuronyl epimerase; hst-2, HS 2-O-sulfotransferase; hst-6, HS 6-O-sulfotransferases; hst-1, N-deacetylase/sulfotransferase; hst-3.1 and hst-3.2, HS 3-O-sulfotransferases (in red). (B) Phylogenetic clustering of HS 3-O-sulfotransferases with ClustalW into two subgroups. Accession numbers: HsHS3ST1 (NP_005105), HsHS3ST5 (NP_705840), DmHs3stA (NP_788409), DmHs3stB (NP_573370), HsHS3ST4 (Q9Y661), HsHS3ST2 (NP_006034), HsHS3ST3B (NP_006032), HsHS3ST3A (NP_006033), CeHST-3.1 (CCD66155), and CeHST-3.2 (NP_001024698). Proteins in bold have a predicted transmembrane domain using a hidden Markov Model (Krogh et al. 2001) at http://www.cbs.dtu.dk/services/TMHMM/. There is no obvious correlation between subgroups and predicted transmembrane domains. (C) Gene structure of hst-3.1 on chromosome II. Based on cDNA analyses, the tm734 allele results in an in frame deletion of 88 amino acids and is a predicted strong loss of function allele (Figure 2). Based on PCR analyses and sequencing, the ok1249 allele results in an approximate 0.9kb deletion of the promoter leaving at least 125 bp upstream of the hst-3.1 transcription start site. The hst-3.1 transcript is transpliced to SL1. In panels C and D, exons shaded in red encode the sulfotransferase domains and 5′- and 3′-PAPS binding sites are indicated in blue. (D) Gene structure of hst-3.2 on chromosome X. The hst-3.2 transcript is transpliced to SL1. The position of deletion and point mutant alleles are shown. Based on cDNA analyses, the hst-3.2(tm3006) allele results in a frameshift after 121 of 291 amino acids and a premature stop after two additional nonconserved amino acids (Figure 2B) whereas hst-3.2(tm3208) creates a frameshift after 175 of 291 amino acids with a premature stop after nine nonconserved amino acids (Figure 2B). This leaves a structural determinant of the enzyme partially intact, namely a loop that is predicted to partake in the formation of the groove in which the HS substrate binds (Figure 2, C−E) (Edavettal et al. 2004). Point mutant alleles were isolated by expanding a screen described previously (Bülow et al. 2002) and result in stop codons after 49 amino acids (dz171), 138 amino acids (dz164), or a N51K missense mutation (dz169) which introduces a positive charge in the cosubstrate binding site (Figure 2B). (E) DIC images of control, hst-3.1, hst-3.2, and double mutants. Both single and double mutants of hst-3.1 and hst-3.2 are viable, fertile and display no obvious morphological defects. Scale bar indicates 100 µm.