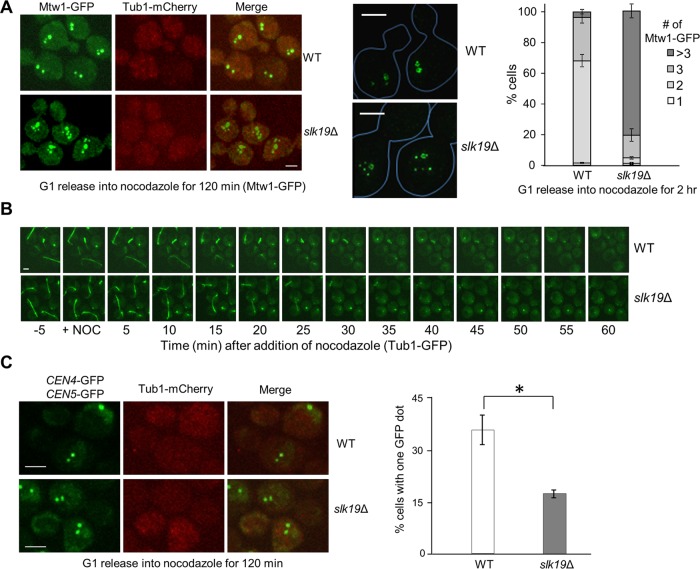
FIGURE 2:
Slk19 is required for kinetochore clustering after nocodazole treatment. (A) slk19Δ mutants show a kinetochore clustering defect. G1-arrested WT and slk19Δ cells in a MTW1-GFP TUB1-mCherry background were released into 20 μg/ml nocodazole for 120 min. After fixation, >100 cells were counted for the number of Mtw1-GFP foci per cell. Right, the average of three separate experiments. Left, representative images for Mtw1-GFP and Tub1-mCherry signals from confocal microscopy. Middle, representative images for Mtw1-GFP signals from superresolution microscopy. Bar, 3 μm. (B) slk19Δ mutant cells show similar microtubule-depolymerizing kinetics as WT cells. WT and slk19Δ cells with Tub1-GFP were loaded into a chamber where YPD medium containing 20 μg/ml nocodazole started to flow over the cells at time 0. Maximum-projection images were then created to show the spindle morphology over time. (C) Fewer slk19Δ cells show colocalized centromeres from chromosomes IV and V after nocodazole treatment. WT and slk19Δ cells in a TUB1-mCherry background with GFP-marked centromeres of chromosome IV and V (CEN4-GFP CEN5-GFP) were grown to log phase at 25°C. After G1 arrest, the cells were released into 20 μg/ml nocodazole for 120 min and then harvested and fixed for fluorescence microscopy. The number of GFP dots per cell was counted in >100 cells, and the average from triplicates is shown. Confocal microscopy was used to project representative maximum-intensity images for CEN4-GFP, CEN5-GFP, and Tub1-mCherry. Bar, 3 μm.