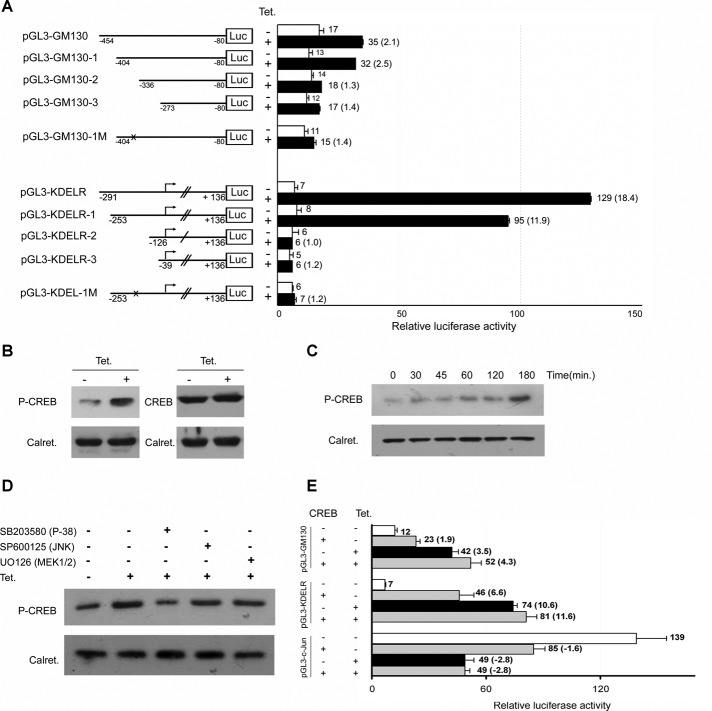
FIGURE 6:
CREB activation and its consensus sequence are required for the effect of Rab1b. (A) Relative luciferase activities obtained with a set of reporter vectors containing the indicated 5′-flanking variants of the GM130 and KDELR promoters depicted in Figure 3A. pGL3-GM130-1/2/3 and pGL3-KDELR-1/2/3 represent different deletions, and pGL3-GM130-1M and pGL3-KDELR-1M represent mutant variants of their respective versions in which the CREB-binding consensus site was mutated. The graphs show relative luciferase activities; numbers on the right of each bar indicate the average value of relative luciferase activity from at least three experiments and the ratio between samples for the indicated condition is shown in parentheses. Error bars represent SD. (B–D) Western blot analysis of T-Rex Rab1b cells. (B) Detection of P-CREB and total CREB in the absence or presence of tetracycline (over 48 h). (C) Detection of P-CREB at the indicated times after tetracycline addition. (D) Effect of the indicated MAPK inhibitors on the CREB phosphorylation induced by an increase in Rab1b levels. Calreticulin was used as the loading control. (E) Luciferase activities obtained with the indicated pGL3 constructs (represented in Figure 4A) cotransfected with a plasmid vector encoding CREB and with a control vector (R. reiniformis phRL-TK) in the T-Rex Rab1b cells. CREB expression was able to stimulate GM130 and KDELR transcription and to inhibit c-Jun transcription. While these modifications were less pronounced than those observed by an increase in Rab1b, they were still significant (p < 0.05) and indicated that CREB can regulate their promoters. Additionally, modifications in the promoter activities in cells expressing both CREB and Rab1b had no significant differences compared with those induced by Rab1b alone.