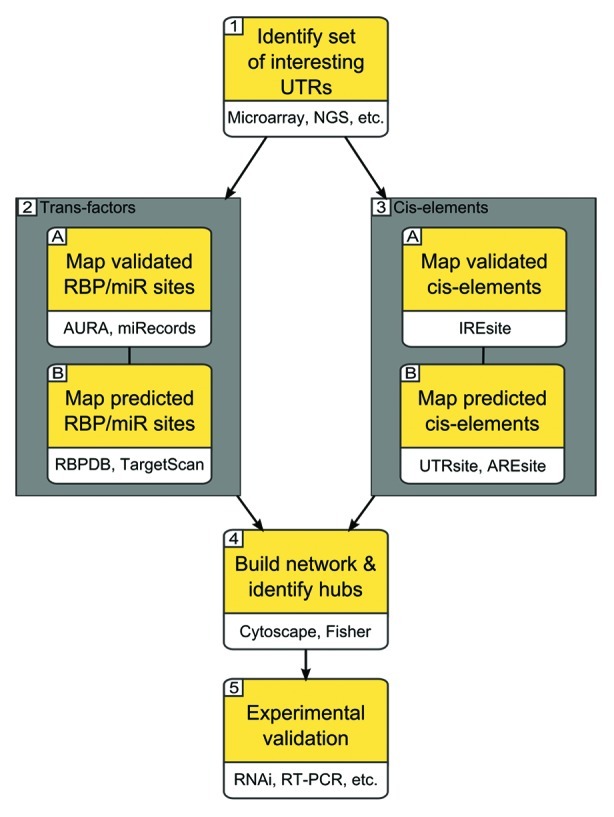
Figure 2. A possible discovery pipeline for post-transcriptional regulatory mechanisms. The workflow starts by the selection of interesting UTRs: these may, for instance, come from high-throughput experiments done by the microarrays or next generation sequencing technologies. The pipeline then proceeds by searching for both experimentally validated and computationally predicted trans-factors binding sites and cis-elements over these UTRs. The resulting interactions are then collected into a network: important nodes are identified by enrichment tests such as the Fisher test. Interesting leads are eventually subjected to experimental validation by various methods of targeted gene expression perturbation, as RNA interference.
