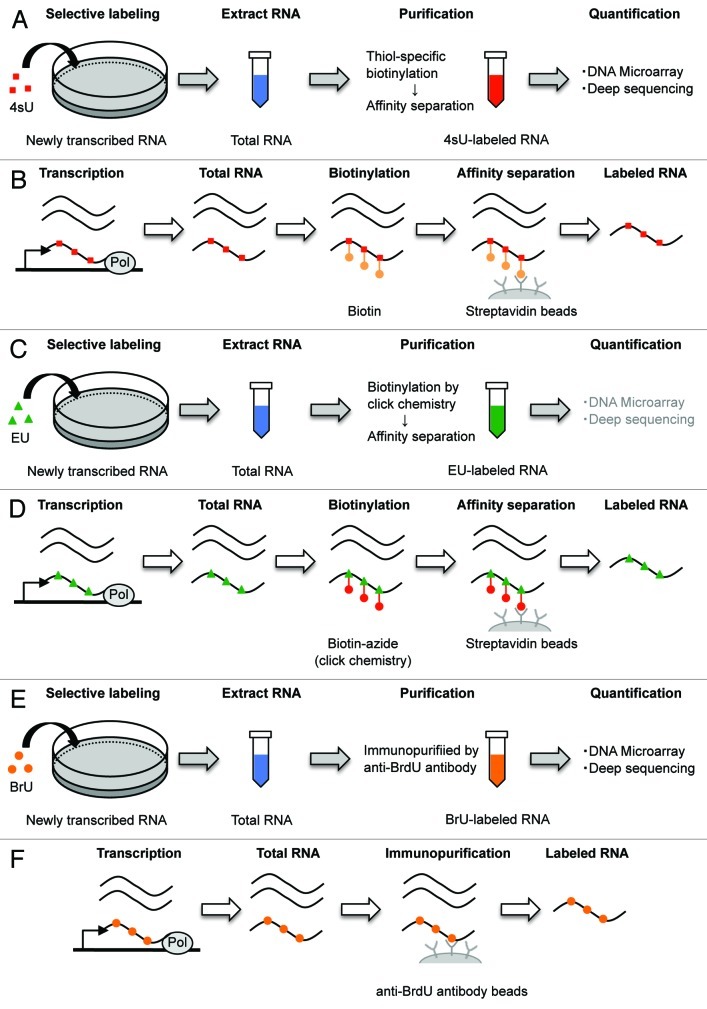
Figure 2. Schematic representation of the genome-wide method for RNA decay using pulse labeling with uridine analogs. (A) 4sU is added to cells for a pre-defined time. Collecting the cells and extract total RNA (blue). Then, 4sU-labeled RNAs (red) are isolated by thiol-specific biotinylation and affinity separation. (B) 4sU is introduced into newly transcribed RNA in place of uridine. Total RNA extract is biotinylated by covalently linking biotin (orange) to 4sU, followed by binding streptavidin coated beads (gray). Biotinylated RNA is isolated, whereas unlabeled RNA is wash out. Then, cleaving the biotin-4sU disulfide bond releases the 4sU-labeled RNA from beads. (C) EU is added to cells for a pre-defined time. Collecting the cells and extract total RNA (blue). Then, EU-labeled RNAs (green) are isolated by biotinylation of EU in click chemistry and affinity separation. (D) EU is incorporated into newly transcribed RNA in place of uridine. Total RNA extract is biotinylated by copper-catalyzed cycloaddition reaction (click chemistry) (red) to EU, followed by isolation of EU-labeled RNA, similarly to 4sU method. (E) BrU is added to cells for a pre-defined time. Collecting the cells and extract total RNA (blue) at sequential time points after removal of surplus BrU from the culture medium. Then, BrU-labeled RNAs (orange) are isolated by immunopurification. (F) BrU is incorporated into newly transcribed RNA in place of uridine. BrU-labeled RNAs in total RNA extract are immunopurified using an anti-BrdU antibody coated beads (gray), which recognizes both BrdU and BrU.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.