Abstract
The recent identification of the oxygen-sensing mechanism in plants is a breakthrough in plant physiology. The presence of a conserved N-terminal motif on some ethylene responsive factors (ERFs), targets the protein for post-translational modifications finally leading to degradation under normoxia and thus providing a mechanism for sensing the presence of oxygen. The stabilization of the N-terminus under low oxygen activates these ERFs, which regulate low oxygen core genes that enable plants to tolerate abiotic stress such as flooding. Additional mechanisms that signal low-oxygen probably also exist, and the production of reactive oxygen species (ROS) has been observed under low oxygen, suggesting that ROS might be part of the network involved in plant acclimation. Here, we review the most recent findings related to oxygen sensing.
Keywords: ROS, anoxia, hypoxia, low oxygen
Plants are inevitably subject to low oxygen (O2), mainly due to the metabolic requirement of cells with high O2 demand or reduced availability, such as in bulky storage organs, seeds, meristem tissues and fruits.1 Climatic changes have exacerbated the shortage of O2, with more frequent excess rainfall leading to plant submergence in flood-prone river areas and farmlands. Dramatic flooding often affects marginal lands where food demand is high, thus tolerant varieties are crucial in order to increase crop yield.
Decline in O2 availability results in a reduction in the energy produced by the plant, because of the shift from aerobic to anaerobic respiration, which is less efficient in terms of ATP synthesis. A limited reduction in O2 (hypoxia) results in a reorganization of metabolic fluxes, so that energy usage is optimized to fulfill the house-keeping activities of the cell which are required to prevent disorganization and death.2,3 The complete absence of O2 (anoxia) is considerably more problematic for cells, which under such conditions can only rely on glycolysis for ATP production, thus rapidly entering into an energy crisis.4
Many plant species have evolved adaptive mechanisms to survive low O2, which in some cases O2 (anoxia) is led to a tolerance to flooding. The molecular aspects governing these traits have been partially elucidated in both rice and Arabidopsis, and the centrality of ethylene responsive factors (ERFs) has been identified in plant responses to low O2.5-8 However, other signaling pathways are also likely involved in tolerance. The ability to sense the drop in sugar content following submergence and reactive oxygen species (ROS) production under anoxia are believed to be potential candidates for such sensing.9-11 These multiple mechanisms could work in parallel or play a major/minor role depending on the severity of the stress and the plant's developmental stage.
Ethylene is a Central Component in Rice Adaptation to Low Oxygen
In rice, ethylene plays a crucial role under submergence. When subjected to flooding, both deep water rice and submergence-tolerant lowland rice accumulate ethylene. In deep water rice, ethylene promotes gibberellin-dependent elongation, via the activation of the two ERF genes SNORKEL1 (SK1) and SK2. This results in an “escape” strategy to re-establish contact with the air.6,12 In fact, submergence-induced elongation is one of the most widespread escape mechanisms in plants, with Rumex palustris providing an excellent example.13-15 However, this strategy is only successful if enough energy is available throughout the whole period of submergence.
Submergence-tolerant lowland rice adopt the opposite strategy: gibberellin signaling is repressed, thus inhibiting growth, through the ERF gene SUBMERGENCE 1A (SUB1A) and the downstream repressors of gibberellin action, namely SLENDER1 (SLR1) and SLENDER-LIKE1 (SLRL1). This results in a “quiescence” strategy that reduces energy consumption during stress.5,16
Ethylene is therefore a central regulator of two opposing strategies to survive different habitats. Both these regulative pathways are realized through the action of ERF genes belonging to group VII, reinforcing the hypothesis that diverging ERFs can provide distinct tolerance mechanisms in multiple species.1
Direct Low Oxygen Sensing in Arabidopsis
Animals are known to have a direct O2 sensing mechanism since several years, while until recently a plant-counterpart of this sensing mechanism remained elusive.17,18 In animals, a transcription factor (TF), namely the hypoxia-inducible factor 1 α (HIF1α) is the target of proteasomal degradation via O2-dependent hydroxylation under normoxia. Oxygen depletion strongly inhibits HIF-1α degradation, with downstream induction of hypoxia-responsive genes.19 A similar mechanism was thought to be present in plants too, but no orthologs of HIF-1α have been found.
Recently, two independent studies suggested that the N-end rule pathway (NERP) for protein degradation might be involved in the regulation of the low O2 response in plants.7,8 The NERP recognizes a specific N-terminus motif for post-translational modification, leading finally to the (de)stabilization of the protein.20 A conserved amino acid motif corresponding to Met-Cys was found in ERFs of group VII, which had previously been shown to be involved in low O2 genes regulation.21,22 In fact, a MetCys Nend has been found in HRE1, HRE2, RAP2.2 and RAP2.12 of Arabidopsis, but also in the rice proteins SUB1A and SK1/SK2, responsible for “quiescence” and “escape” strategy, respectively, for tolerance in Oryza sativa. The Cys present at the N-terminus of these proteins is oxidized in the presence of O2 and thus leads to post-translational modification ending with their degradation.7,8 However, under low O2, the N-terminus is stabilized.
Rice SUB1A-1 regulation has been shown to be uncoupled from the N-end rule pathway, likely due to the absence of a crucially positioned lysine downstream of the N-end or to differences in the tertiary structure.7 The higher stability of SUB1A may explain the tolerance of SUB1A-containing rice varieties to abiotic stresses irrespectively of low O2 availability.7
Licausi and colleagues identified one specific ERF protein, RAP2.12, which is believed to act as an O2 sensor in plants. RAP2.12 is expressed constitutively and ubiquitously, and the RAP2.12 protein, which would otherwise be degraded as a consequence of Cys oxidation in normoxia, is protected from degradation by docking Acyl-CoA-Binding Proteins ACBP1 and ACBP2, which are located in the plasma membrane.8 Under hypoxia, RAP2.12 is released from the plasma membrane and migrates to the nucleus, thus activating downstream events for hypoxia tolerance.8 The NERP mutants ate1ate2 and prt6, which are defective in steps related to proteasomal degradation, revealed the constitutive expression of hypoxic core genes such as ADH1, SUS4 and PDC1.7 In addition, plants carrying a stabilized HRE2, through the modification of the N-terminus, showed an enhanced survival under hypoxia.7
The identification of an O2 sensing mechanism in plants is a breakthrough in plant science which opens up important questions, the most critical being the mechanism of Cys oxidation and the regulation of downstream signaling that mediates variations in hypoxia tolerance (Fig. 1).23
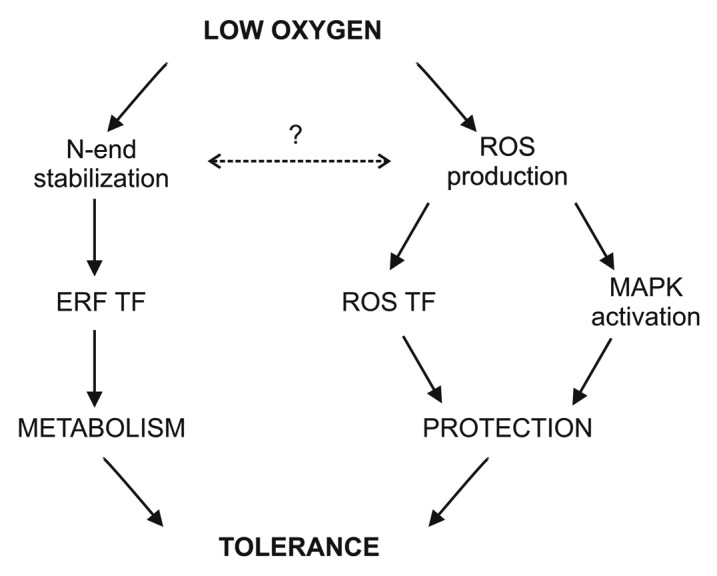
Figure 1. Arabidopsis signal transduction events under low O2. Under hypoxia and anoxia, the N-end of some group VII ERFs is stabilized thus activating anaerobic core genes. Under anoxia, a mitochondrial imbalance produces ROS that regulate MAPKs. A ROS rheostat mediated by NADPH oxidase is also activated and TFs related to ROS signaling are induced, thus promoting ROS-related genes. Both these processes help plants to be tolerant to low O2.
Indirect Low Oxygen-Sensing in Arabidopsis Involves ROS
The depletion of O2 has rapid consequences at a molecular level, as is demonstrated by several experimental microarray analyses performed on plants in which both the O2 levels and length of O2 deprivation was varied.22,24-27 Some meta-analyses identified low O2 core genes.2,29 It was thus evident that although several genes modulated by low O2 are related to anaerobic metabolism, this set of genes also includes many proteins with an unknown function and oxidative stress-related genes.2,24 Heat shock proteins (HSPs) were found to be a conserved group of low-oxygen-responsive genes in different kingdoms and the overexpression of the transcription factor HsFA2 was shown to increase anoxia tolerance, irrespectively of the induction of anaerobic genes.24,29
In fact, adh- plants do cross-acclimate to anoxia following a mild heat-stress treatment, thus indicating that the effects of heat on anoxia tolerance are unrelated to the fermentative metabolism.30 This would imply a tolerance mechanism that does not rely on low O2 metabolic acclimation alone, but which is linked to cell protection from stress. This phenomenon is likely in common with heat stress, which has been shown to induce cross protection to anoxia.30 HsfA2 has been suggested to be a sensor of H2O2 presence and its expression is observed in several systems producing ROS.31,32 In addition, HSP production as a response to H2O2 is conserved in several kingdoms.33 A ROS-dependent acclimation could work in parallel to metabolic acclimation and could be critical should the latter fail. A link between ROS production and acclimation to hypoxia was proposed by Baxter-Burrel and colleagues, who suggested the existence of an anaerobic H2O2 production mediated by a NADPH oxidase (Rboh) regulated by a ROP/ROPGAP rheostat.34 RBOHD has been found to be positively regulated by both low O2 and ROS.11,35 This NADPH isoform is transcriptionally responsive to many abiotic stresses.35 Interestingly, rbohD- plants are less tolerant to anoxia tolerance than wild types.11 Indeed, HsfA2 and ZAT12 expression display a lower induction in rbohD- under anoxia, possibly explaining the lower tolerance.11
The presence of an oxidative burst under low O2 has also been proposed by Banti and colleagues, who linked heat-anoxia cross-tolerance to H2O2 production, a common element under both heat and anoxia.24 Besides H2O2 production mediated by NADPH oxidase(s), an alternative (or additional) possible source of ROS under anoxia may be mETC.10 Anoxic treatment induces ROS production in mitochondria, probably due to ETC impairment by O2 depletion.10 A downstream positive regulation of MPK3, 4 and 6 seems to be dependent on this oxidative burst. In Chang's study, overexpression on MPK6 was responsible for improved tolerance to anoxia but anaerobic core genes were not markedly altered, neither were they in mpk6 plants.10
The signaling pathway that modulates the expression of HSPs under anoxia is likely distinct from the one modulating metabolic acclimation to this stress. Anaerobic core genes are ectopically activated in N-end rule mutants, whereas the group of HSPs commonly regulated in several low O2 experiments are not.11 HSPs are significantly induced under anoxia but not hypoxia, suggesting that the response to hypoxia and anoxia is modulated by a common signaling pathway for metabolic adaptation alone. Severe low O2 conditions (such as anoxia) seem to activate a signaling pathway that includes ROS production. Although ROS could be produced under anoxia as a general stress, the transient and rapid peak of H2O2 observed soon after the beginning of an anoxic treatment indicates a possibly specific mechanism to modulate ROS production.10,11
Conclusions
Recent results have unveiled the presence of distinct signaling mechanisms that modulate plant responses to low O2. Together with a master regulation occurring through the stabilization of the N-terminus of some group VII ERF proteins, ROS production under anoxia likely represents an important component of the anaerobic signaling network in plants. While NERP-mediated transcriptional regulation may take place irrespectively of level of severity of the stress, ROS production and its signaling cascade are probably related to severe conditions of O2 deprivation only, such as anoxia. Although the production of ROS under anoxia may be paradoxical, it has been shown to occur very early and to be transient, thus not persisting under anoxia.10,11 ROS-related genes have been found to be systematically modulated by anoxia, but the cellular localization of ROS production is still a matter of debate. Recent results support both the mETC and NADPH-oxidases located at the plasma-membrane as possible sources. Two intriguing open issues are (1) understanding how distinct sources of ROS under anoxia might have distinct roles in the way plants acclimatize to anoxia, and (2) whether O2 sensing through the N-end rule is affected by ROS produced at the onset of anoxia.
Acknowledgments
This work was supported by the Scuola Superiore Sant’Anna.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/20322
References
- 1.Bailey-Serres J, Voesenek LACJ. Life in the balance: a signaling network controlling survival of flooding. Curr Opin Plant Biol. 2010;13:489–94. doi: 10.1016/j.pbi.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:18843–8. doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rocha M, Licausi F, Araújo WL, Nunes-Nesi A, Sodek L, Fernie AR, et al. Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol. 2010;152:1501–13. doi: 10.1104/pp.109.150045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Licausi F. Regulation of the molecular response to oxygen limitations in plants. New Phytol. 2011;190:550–5. doi: 10.1111/j.1469-8137.2010.03562.x. [DOI] [PubMed] [Google Scholar]
- 5.Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–8. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- 6.Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009;460:1026–30. doi: 10.1038/nature08258. [DOI] [PubMed] [Google Scholar]
- 7.Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature. 2011;479:415–8. doi: 10.1038/nature10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LA, et al. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature. 2011;479:419–22. doi: 10.1038/nature10536. [DOI] [PubMed] [Google Scholar]
- 9.Lee K-W, Chen P-W, Lu C-A, Chen S, Ho T-HD, Yu S-M. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci Signal. 2009;2:ra61. doi: 10.1126/scisignal.2000333. [DOI] [PubMed] [Google Scholar]
- 10.Chang R, Jang CJH, Branco-Price C, Nghiem P, Bailey-Serres J. Transient MPK6 activation in response to oxygen deprivation and reoxygenation is mediated by mitochondria and aids seedling survival in Arabidopsis. Plant Mol Biol. 2012;78:109–22. doi: 10.1007/s11103-011-9850-5. [DOI] [PubMed] [Google Scholar]
- 11.Pucciariello C, Parlanti S, Banti V, Novi G, Perata P. Reactive oxygen species-driven transcription in Arabidopsis under oxygen deprivation. Plant Physiol. 2012;159:184–96. doi: 10.1104/pp.111.191122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagai K, Hattori Y, Ashikari M. Stunt or elongate? Two opposite strategies by which rice adapts to floods. J Plant Res. 2010;123:303–9. doi: 10.1007/s10265-010-0332-7. [DOI] [PubMed] [Google Scholar]
- 13.Jackson MB. Ethylene-promoted elongation: an adaptation to submergence stress. Ann Bot. 2008;101:229–48. doi: 10.1093/aob/mcm237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heydarian Z, Sasidharan R, Cox MC, Pierik R, Voesenek LA, Peeters AJ. A kinetic analysis of hyponastic growth and petiole elongation upon ethylene exposure in Rumex palustris. Ann Bot. 2010;106:429–35. doi: 10.1093/aob/mcq138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voesenek LACJ, Benschop JJ, Bou J, Cox MC, Groeneveld HW, Millenaar FF, et al. Interactions between plant hormones regulate submergence-induced shoot elongation in the flooding-tolerant dicot Rumex palustris. Ann Bot. 2003;91(Spec No):205–11. doi: 10.1093/aob/mcf116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey-Serres J, Voesenek LACJ. Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol. 2008;59:313–39. doi: 10.1146/annurev.arplant.59.032607.092752. [DOI] [PubMed] [Google Scholar]
- 17.Goldberg MA, Dunning SP, Bunn HF. Regulation of the erythropoietin gene: evidence that the oxygen sensor is a heme protein. Science. 1988;242:1412–5. doi: 10.1126/science.2849206. [DOI] [PubMed] [Google Scholar]
- 18.Semenza GL, Nejfelt MK, Chi SM, Antonarakis SE. Hypoxia-inducible nuclear factors bind to an enhancer element located 3′ to the human erythropoietin gene. Proc Natl Acad Sci U S A. 1991;88:5680–4. doi: 10.1073/pnas.88.13.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci U S A. 1993;90:4304–8. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–86. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 21.Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, et al. Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010;153:757–72. doi: 10.1104/pp.110.155077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, et al. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 2010;62:302–15. doi: 10.1111/j.1365-313X.2010.04149.x. [DOI] [PubMed] [Google Scholar]
- 23.Sasidharan R, Mustroph A. Plant oxygen sensing is mediated by the N-end rule pathway: a milestone in plant anaerobiosis. Plant Cell. 2011;23:4173–83. doi: 10.1105/tpc.111.093880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banti V, Mafessoni F, Loreti E, Alpi A, Perata P. The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 2010;152:1471–83. doi: 10.1104/pp.109.149815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J. Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann Bot. 2005;96:647–60. doi: 10.1093/aob/mci217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loreti E, Poggi A, Novi G, Alpi A, Perata P. A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 2005;137:1130–8. doi: 10.1104/pp.104.057299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dongen JT, Fröhlich A, Ramírez-Aguilar SJ, Schauer N, Fernie AR, Erban A, et al. Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of arabidopsis plants. Ann Bot. 2009;103:269–80. doi: 10.1093/aob/mcn126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, et al. Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol. 2011;190:457–71. doi: 10.1111/j.1469-8137.2010.03590.x. [DOI] [PubMed] [Google Scholar]
- 29.Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, et al. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 2010;152:1484–500. doi: 10.1104/pp.109.151845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banti V, Loreti E, Novi G, Santaniello A, Alpi A, Perata P. Heat acclimation and cross-tolerance against anoxia in Arabidopsis. Plant Cell Environ. 2008;31:1029–37. doi: 10.1111/j.1365-3040.2008.01816.x. [DOI] [PubMed] [Google Scholar]
- 31.Miller G, Mittler R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot. 2006;98:279–88. doi: 10.1093/aob/mcl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, et al. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;141:436–45. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandenbroucke K, Robbens S, Vandepoele K, Inzé D, Van de Peer Y, Van Breusegem F. Hydrogen peroxide-induced gene expression across kingdoms: a comparative analysis. Mol Biol Evol. 2008;25:507–16. doi: 10.1093/molbev/msm276. [DOI] [PubMed] [Google Scholar]
- 34.Baxter-Burrell A, Yang Z, Springer PS, Bailey-Serres J. RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science. 2002;296:2026–8. doi: 10.1126/science.1071505. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol. 2011;14:1–9. doi: 10.1016/j.pbi.2011.07.014. [DOI] [PubMed] [Google Scholar]


