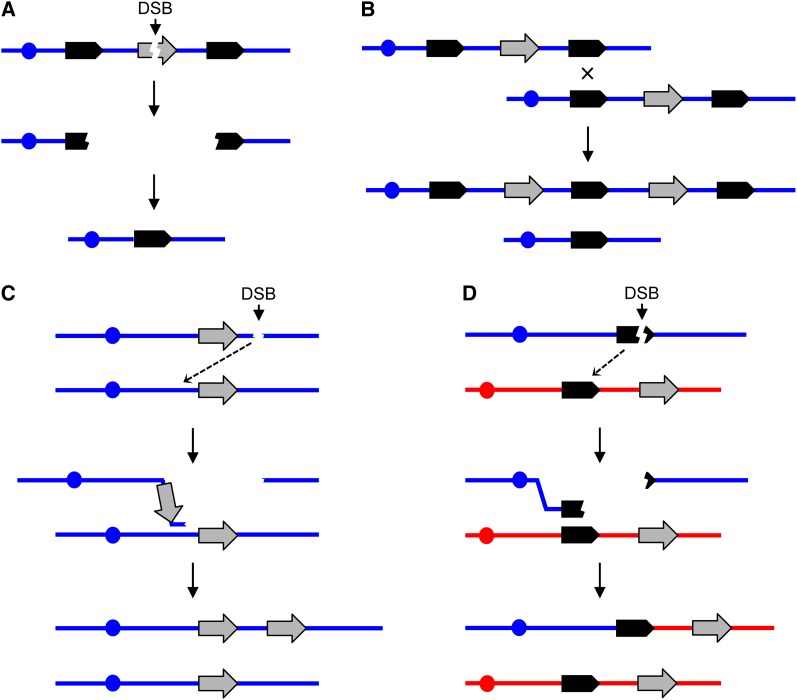
Figure 1.
General mechanisms for the generation of deletions and duplications. In this figure, chromosomes are shown as horizontal blue or red lines, DNA repeats as solid arrows, reporter genes as shaded arrows, and centromeres as circles. Single DNA strands are not represented. Variations of these models are also possible. (A) Deletion formation by the single-strand annealing (SSA) pathway. A double-strand DNA break (DSB) between the flanking repeats results in two broken ends that are resected 5′ to 3′. When complementary single-strand regions of the flanking repeats are exposed, reannealing occurs, resulting in loss of the sequences between the repeats (Paques and Haber 1999). (B) Unequal crossovers resulting in deletions and duplications. This homologous recombination event could involve either sister chromatids in haploids or diploids or homologous chromosomes in diploids. (C) Microhomology-mediated replication. A free 3′ end associated with centromere-containing DNA fragment invades a sister chromatid or a homologous chromosome using a nonallelic microhomology sequence. The subsequent break-induced replication (BIR) event generates a duplication in which the breakpoints share little sequence homology. It is assumed that the DNA fragment without a centromere is lost. (D) Coupled terminal duplication and deletion resulting in a nonreciprocal translocation. The different homologs are shown in blue and red. A DSB in or distal to a repeat in the blue chromosome is repaired by a BIR event using a repeat on a nonhomologous chromosome (red). This event results in a duplication of sequences from the red chromosome and a deletion of sequences from the blue chromosome.