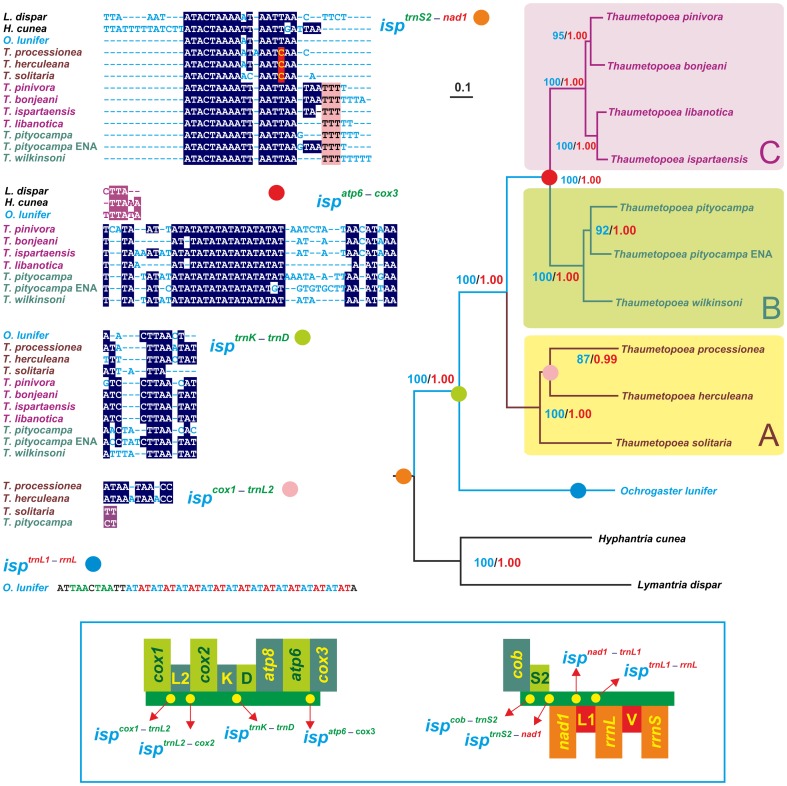
Figure 4. The reference phylogenetic tree and evolution of mitochondrial intergenic spacers.
Maximum likelihood tree (−ln = 44305.147421) inferred from the aag13sp-set ALN. The analysis was performed by applying the GTR+G evolutionary model and according to the most complex partitioning scheme described in the main text. Blue-coloured numbers indicate bootstrap values expressed as percentage, whereas red-coloured numbers indicate posterior probabilities computed through Bayesian inference analysis on the same data set. The scale bar represents 0.1 substitutions/site. Occurrence and evolutionary pathway of isps exhibiting a uniqueness in term of genomic position plus sequence identity. Multiple alignments of isps, representing unique molecular signatures, are provided with invariant positions depicted on a blue background. Red/pink background is used to identify nucleotides characterizing peculiar clades. Single sequences/multiple alignments of isps, not representing unique molecular signatures, are depicted on a purple background. The inset on the bottom shows the placement of isps in the lepidopteran complete mitochondrial genome. Genes are coloured as in Figure 1.