Figure 9.
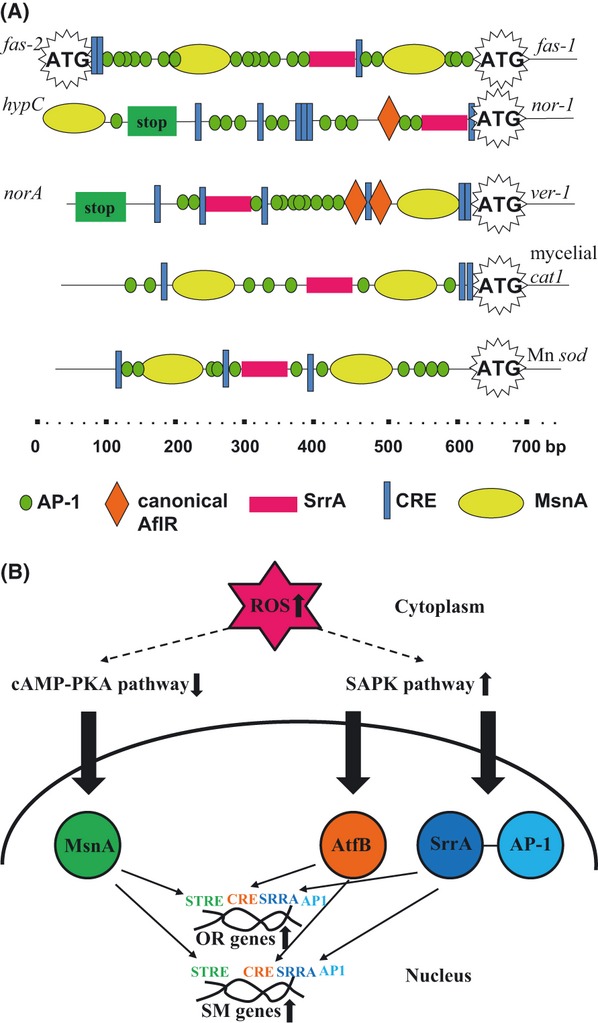
Proposed model for transcriptional activation of secondary metabolism and oxidative stress-response genes by binding of transcription factors in response to oxidative stress. (A) Schematic representation of the transcription factor-binding sites in the promoters of the aflatoxin biosynthetic and antioxidant genes used in this study. (B) Regulatory network for transcriptional activation of secondary metabolism and oxidative stress-response genes for cellular defense against oxidative stress. Based on available experimental evidence, we propose that increased levels of intracellular ROS in fungal cells downregulate the cAMP-PKA signaling pathway. This promotes MsnA binding to stress-response element (STRE) sites in promoters of oxidative stress-response (OR) genes including antioxidant genes for their activation. Simultaneously, ROS upregulates the SAPK signaling cascades. This promotes AtfB and SrrA binding (SrrA recruits AP-1) to corresponding CRE, SRRA, and AP1 sites in promoters of the oxidative stress-response (OR) genes including the antioxidant genes for their induction. Then, MsnA, AtfB, and SrrA bind (SrrA recruits AP-1) to corresponding STRE, CRE, SRRA, and AP1 sites in promoters of secondary metabolism genes including aflatoxin genes for their activation due to excess ROS. Secondary metabolites produced in response to ROS play a role in a defense mechanism of fungal cells against oxidative stress. PKA, protein kinase A; SAPK, stress-activated protein kinase; OR, oxidative stress response; SM, secondary metabolism.
