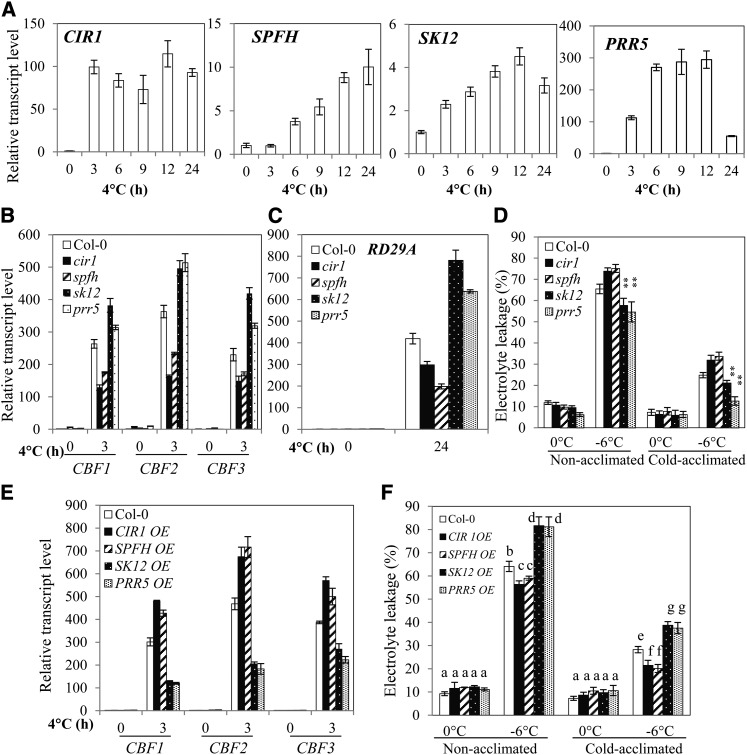
Figure 5.
CIR1, SPFH, SK12, and PRR5 Regulate Expression of CBF Genes and Cold Tolerance.
(A) CIR1, SPFH, SK12, and PRR5 are cold inducible in the wild type.
(B) and (C) Relative transcript levels of CBF1, CBF2, CBF3, and RD29A in cir1, spfh, sk12, and prr5 mutant plants.
(D) Freezing tolerance of cir1, spfh, atsk12, and prr5 mutant plants determined by electrolyte leakage assays. **P < 0.01, as determined by Student’s t test.
(E) Relative transcript levels of CBF1, CBF2, and CBF3 in overexpression plants of CIR1 (CIR1 OE), SPFH (SPFH OE), At-SK12 (SK12 OE), and PRR5 (PRR5 OE).
(F) Freezing tolerance of CIR1 OE, SPFH OE, AtSK12 OE, and PRR5 OE plants. Cold acclimation in (D) and (F) was achieved by incubating plants at 4°C for 1 week. Data in (E) and (F) are from one representative transgenic line of two independent transgenic lines of CIR1 OE, SPFH OE, At-SK12 OE, and PRR5 OE plants (see Supplemental Figures 11E and 11F online).
Error bars indicate the sd (n = 4 in [B], [C], and [E]; n = 12 to 15 in [D] and [F]). One-way ANOVA (Tukey-Kramer test) was performed for data in (F), and statistically significant differences are indicated by different lowercase letters (P < 0.01). Experiments in were repeated at least four times with similar results, and values shown are from one experimental repetition.