Figure 1.
Functional Proteins with Different Proportions of Intrinsic Disorder.
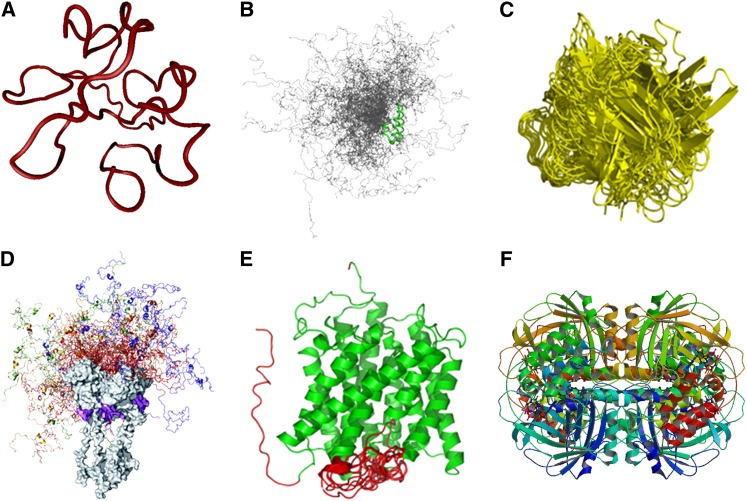
(A) Random coil. The structure shown is one of the 10 conformers in an ensemble of solution NMR structure of HIV-1 Tat protein (Protein Data Bank code: 1TIV).
(B) Premolten globule, which is more compact than random coil but still mostly disordered with some local residual secondary structures. The example shown is the conformational ensemble containing 200 conformers of the natively disordered region of PX, the nucleocapsid binding domain of Sendai virus phosphoprotein (Bernadó et al., 2005); the regions in green indicate the local ordered structures along the flexible chain of PX.
(C) Molten globular state, which retains all secondary structures and compact shape with the side chains changing from rigid to nonrigid packing. This state is responsible for biological functions such as translocation of proteins across membranes. The example shown is the unstable conformers of Allergen PHL P2 (Protein Data Bank code: 1WHO) generated from molecular dynamic simulations during interaction with receptor (Liang et al., 2009).
(D) Protein with ordered domain and premolten globule-like domain. The example shown is full-length human tumor repressor p53 tetramer-DNA complex; the C-terminal ordered domains (gray) and DNA (magenta) are shown in space-fill mode. The conformational ensemble of the disordered N-terminal domains from the four different monomers is shown in different colors, and 20 conformers are shown for each monomer (Wells et al., 2008).
(E) Protein with ordered domain and random coil. The example shown is an integral membrane protein involved in forming an ammonia channel (Protein Data Bank code: 2NMR); the helical bundle is shown in green and the disordered regions in red (Xue et al., 2009).
(F) Ordered protein. The example shown is the crystal structure of beef liver catalase with an NADPH binding site (Protein Data Bank code: 8CAT).