Fig 8.
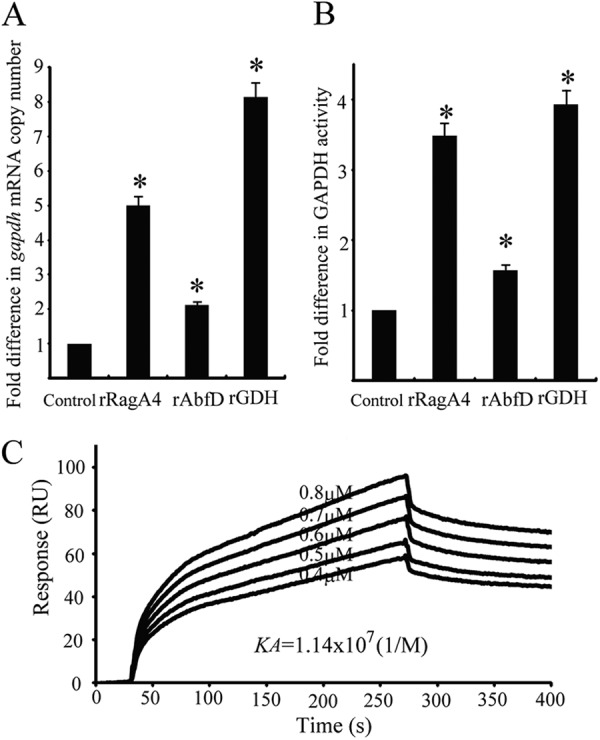
Effect of P. gingivalis client proteins on expression of P. gingivalis GAPDH. (A) gapdh mRNA in P. gingivalis was measured by quantitative RT-PCR. Transcript levels were normalized to 16S rRNA. P. gingivalis without recombinant proteins served as a control. The fold difference is expressed as gapdh mRNA levels after exposure to P. gingivalis client proteins (5 μg) compared to the control. Data are means and standard deviations from three independent experiments performed in triplicate. An asterisk denotes significant difference at P < 0.01 (t test) compared to P. gingivalis alone. (B) Measurement of P. gingivalis cell surface GAPDH activity. GAPDH activity was measured at 340 nm. The fold difference is presented as a ratio of GAPDH activity with P. gingivalis client proteins (5 μg) to that without the client proteins. P. gingivalis without recombinant protein served as a control. Data are means and standard deviations from three independent experiments performed in triplicate. An asterisk denotes a significant difference at P < 0.01 (t test) compared to P. gingivalis alone. (C) Sensorgrams of P. gingivalis rFimA binding to immobilized P. gingivalis rGAPDH in kinetic studies. P. gingivalis rFimA fimbriae were injected over P. gingivalis rGAPDH on the sensor chip at various concentrations (0.4 to 0.8 μM).
