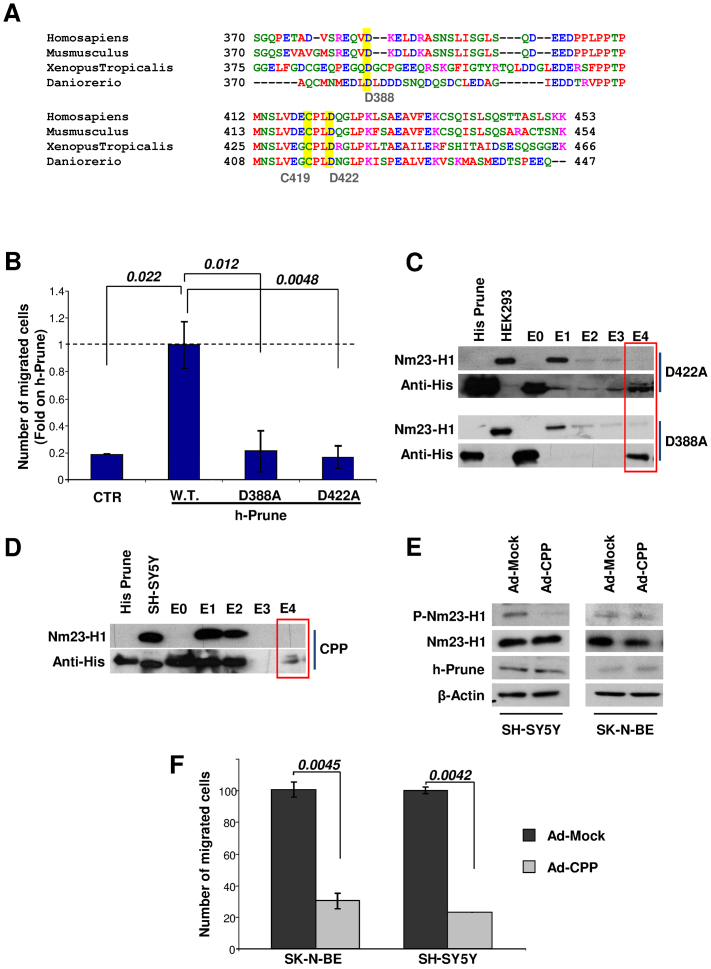
Figure 4. Migration assays to assert migration properties of differential protein domains of the h-Prune protein.
(a) Alignment of the h-Prune C-terminal. Amino acids involved in Nm23-H1 binding are highlighted in yellow. (b) Two-dimensional invasion assay. HEK293 cells transfected with h-Prune mutant proteins (D388A and D422A) showed decreased migration ability. Data are represented as relative (fold) increases in the number of cells migrating compared to full-length h-Prune wild-type transfected cells. (c) Affinity chromatography. The His-tagged 354-453 h-Prune D422A mutant (upper) and D388A mutant (lower) were immobilized on the resin. HEK293 total extract was loaded as control. An anti-His antibody was used as control for the protein immobilized. An anti-Nm23-H1 antibody shows that the mutated 354–453 h-Prune interacts weakly with Nm23-H1. (d) Affinity chromatography. SH-SY5Y pre-infected cells (Ad-CPP) were loaded onto the chromatography column and the eluates were loaded onto acrylamide gels. Nm23-H1 was detected using an anti-Nm23-H1 antibody. Protein 373–353 h-Prune was revealed using an antibody against the His tag. (e) Following Ad-CPP and Ad-Mock infections in SH-SY5Y and SK-N-BE cells, the protein levels of phospho-Nm23-H1, Nm23-H1 and anti-h-Prune were analyzed by Western blotting. β-Actin was used as the loading control. (f) Two-dimensional migration assay of NBL cells showing that CPP overexpression reduces the migratory properties of both SH-SY5Y and SK-N-BE cells.