Abstract
The nuclear periphery is a dynamic, structured environment, whose precise functions are essential for global processes—from nuclear, to cellular, to organismal. Its main components—the nuclear envelope (NE) with inner and outer nuclear membranes (INM and ONM), nuclear pore complexes (NPC), associated cytoskeletal and nucleoskeletal components as well as chromatin are conserved across eukaryotes (Fig. 1). In metazoans in particular, the structure and functions of nuclear periphery components are intensely researched partly because of their involvement in various human diseases. While far less is known about these in plants, the last few years have seen a significant increase in research activity in this area. Plant biologists are not only catching up with the animal field, but recent findings are pushing our advances in this field globally. In recognition of this developing field, the Annual Society of Experimental Biology Meeting in Salzburg kindly hosted a session co-organized by Katja Graumann and David E. Evans (Oxford Brookes University) highlighting new insights into plant nuclear envelope proteins and their interactions. This session brought together leading researchers with expertise in topics such as epigenetics, meiosis, nuclear pore structure and functions, nucleoskeleton and nuclear envelope composition. An open and friendly exchange of ideas was fundamental to the success of the meeting, which resulted in founding the International Plant Nucleus Consortium. This review highlights new developments in plant nuclear envelope research presented at the conference and their importance for the wider understanding of metazoan, yeast and plant nuclear envelope functions and properties.
Keywords: RanGAP, SUN, chromocentre, nuclear envelope, nuclear pore complex, plamina, telomere
One of the lesser conserved components of the nuclear periphery is the nucleoskeleton framework that underlies the INM and structurally supports the NE. In metazoans this structure is termed the lamina and consists of filamentous lamin proteins.1 While a lamina-like structure has been observed underlying the INM in tobacco BY-2 cells—and termed the plant lamina or plamina—its constituents remain to be identified.2 Despite the absence of lamin sequence homologs in plants, several studies have demonstrated the presence of biochemically, structurally, and functionally similar filamentous nuclear proteins that potentially represent plant versions of lamins.2,3 Compelling evidence for such plant lamins was presented by Eric Richards (Boyce Thompson Institute, USA) and Susana Moreno Diaz de la Espina (Centro Investigaciones Biologicas, Consejo Superior de Investigaciones Cientificas, Spain). Richards reported on genetic analysis of four Arabidopsis CROWDED NUCLEUS (CRWN) (formerly Little Nuclei/LINC) mutants. Double and triple crwn mutants reduce plant size and have smaller nuclei suggesting that these proteins fulfill similar functions to lamins in terms of genome organization and maintaining nuclear size.4 In a complementary study, Moreno Diaz de la Espina used electron immuno-localization to show that the onion Nuclear Matrix Constituent Protein 1 (NMCP1), a homolog of AtCRWNs, localizes to the periphery of the INM. Interestingly this localization appears dependent on the cell’s developmental stage, as there are clear differences between the nuclei in actively dividing or mature root cells. An extensive comparative analysis between lamins and NMCP/CRWN proteins highlight their shared features, such as coiled coils with globular tail and head domains, differential expression patterns dependent on development, localization patterns, and biochemical properties. Together, the genetic, cytological, and structural data now strongly support a current working hypothesis that the plant NMCP/CRWN proteins are functional components of the plant nucleoskeleton, an important conceptual foundation for future structure-function relationships in plant nuclei. It will be highly significant to show whether NMCP/CRWN proteins are present in the filaments of the plamina as observed by Fiserova et al.2
Heterochromatin is known to be associated with the nuclear periphery. Evidence from non-plant systems also suggests that indeed some cases of gene silencing involve anchorage of chromatin to the NE.5 In plants, well-defined heterochromatic structures are referred to as knobs or chromocenters. In Arabidopsis, chromocenters are associated with centromeres, and investigation of their dynamic behavior is beginning to shed new light on the functional significance of nuclear architecture and chromatin structure. Arabidopsis chromosomes each have one chromocenter from which euchromatic loops emanate.6 Paul Fransz (University of Amsterdam, The Netherlands) introduced a link between the nuclear periphery and chromocenters. By precisely defining the substructure of domains within chromocenters, he showed that the 180-bp centromeric repeats were on the NE-adjacent edge of the chromocenters, suggesting a possible mechanism of chromocenter anchoring at the nuclear periphery. In addition Fransz detailed the inverse relationship between chromocenter condensation and nuclei size during seed maturation.7 Further, he examined epigenetic marks and Centromeric Histone 3 (CenH3) localization that revealed an intriguing structure of a chromocenter core with a highly methylated periphery. These findings together with dynamic transitions in the appearance of chromocenters in relation to various stresses highlight new research into connections between chromatin structure, the nuclear periphery, and genomic responses in plants. These are the first examples of how the plant NE interfaces with chromatin structures. How this affects gene silencing and activation which occur at the nuclear periphery in non-plant systems will be a focus of future investigations.
In addition to chromocenters, other well-defined heterochromatic structures associated with the NE are telomeres. In metazoans, telomeres are anchored to the nuclear periphery during mitotic and meiotic divisions and this anchorage is thought to be critical in maintaining telomere integrity.8 Specifically in early to middle prophase I of meiosis, telomeres accumulate at the nuclear periphery, an ancient process thought to facilitate homologous chromosome pairing, synapsis, and recombination.9,10 This telomere cluster at the meiotic NE is referred to as the bouquet, now known to involve telomere anchoring to the NE by interactions with Sad1/UNC84 (SUN) proteins in eukaryotic organisms including plants.10 The SUN proteins have also been shown to be NE anchors of telomeres in mitotic divisions.8 The molecular and functional aspects of SUN-telomere anchorage in plants are only now starting to be unraveled, at least in meiosis. To this extent, Sue Armstrong (University of Birmingham, UK) explored the function of Arabidopsis SUN proteins (AtSUN) in telomere localization to the nuclear periphery. Armstrong showed that SUN proteins are required during meiosis as a knock out of both AtSUN1 and AtSUN2 leads to infertility. Hank Bass (Florida State University, USA) reported on 3D immuno-FISH experiments in maize that revealed a meiotic ‘SUN-belt’ layer (using antibodies to maize ZmSUN1 and ZmSUN2) around the nucleus during early to middle meiotic prophase. Bass also reported on recent analysis of the maize desynaptic (dy) mutant in which a candidate gene, ZmSUN3, was identified as the possible causal mutation.11 This further strengthens the links between telomeres and the NE to affect synapsis and meiotic recombination. Research in both labs is providing first evidence that in plants, similar to yeast and animals, SUN proteins play a central role in meiotic processes.
The ZmSUN3 protein discussed by Bass is part of a newly characterized group of SUN proteins, the mid-SUNs or PM3-type SUNs (plant-prevalent Mid-SUN 3 domain, Fig. 2, ref. 12). This group of proteins is characterized by containing the highly conserved SUN domain at the center of the protein instead of at the C-terminus, as is the case with classical SUN proteins. Other structural differences include the presence of three transmembrane domains in mid-SUN proteins, compared with only one transmembrane domain in the C-terminal SUN proteins (Fig. 2).12 These mid-SUN proteins, which appear prevalent in plants,12 were reported by Katja Graumann (Oxford Brookes University, UK) to also be present in other eukaryotic species. This has recently been supported by the characterization of the S. cerevisiae mid-SUN protein SLP1 (SUN-like protein) and its interaction with the classical C-terminal yeast SUN protein Msp3.13 Findings presented by several participants (Bass, Graumann and Tatout) on the previously uncharacterized mid-SUN proteins provide an example in which the plant field leads research efforts to identify and characterize novel nuclear envelope components. Both Graumann and Christophe Tatout (Blaise Pascal University, France) presented information about the molecular interactions of Arabidopsis SUN proteins.14 Tatout showed that each of AtSUN1 and AtSUN2 can interact with both themselves as C-terminal SUNs and also with mid-SUN proteins. In addition, two further groups of NE associated proteins were identified as SUN interactors by both Tatout and the Graumann and Evans labs, uncovering exciting new research directions in plant NE functions. Graumann’s results also suggest that SUN proteins may interact with CRWN/NMCP proteins, an interaction that is indicative of an important linkage between the INM and the nucleoskeleton, which in metazoans, interestingly, is mediated by SUN-lamin interactions.15
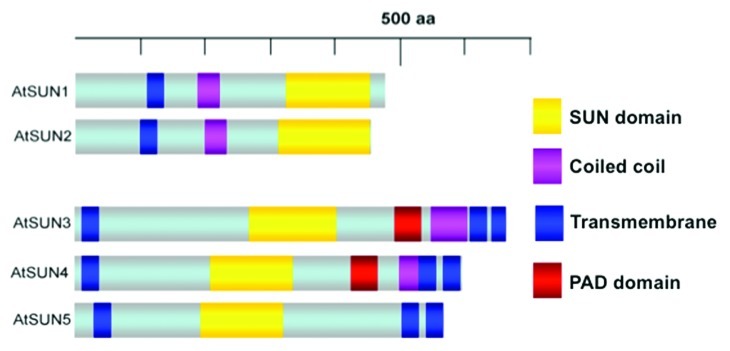
Figure 2. Structural comparison of the two classes of SUN proteins in Arabidopsis. The five SUN proteins present in Arabidopsis thaliana can be sub-divided into two classes: the C-terminal SUN proteins (AtSUN1 and AtSUN2) and the PM3-type or mid-SUN proteins (AtSUN3- AtSUN5). Similar proteins of both sub-groups can also be found in other non-plant eukaryotes. The classical C-terminal SUN proteins contain at least one transmembrane domain and the highly conserved SUN domain at the C-terminus. The PM3-type SUN proteins contain three transmembrane domains, one at the N-terminus and two on the C-terminus while the SUN domain is localized in the middle of the protein. AtSUN3 and AtSUN4 also contain a highly conserved PAD domain (PM3-associated domain), the function of which is currently unknown.12
Iris Meier (The Ohio State University, USA) introduced the first plant nucleo-cytoskeletal bridging complexes. In metazoans and yeast, these complexes consist of INM-localized C-terminal SUN proteins interacting with ONM-localized Klarsicht/Anc-1/Syne-1 homology (KASH) domain proteins, where the SUN proteins link to nucleoskeletal components and the KASH proteins to cytoskeletal elements.15 Their molecular composition and numerous functions, including nuclei movement and positioning, centrosome anchoring, chromosome movement and homologous recombination, are well documented in non-plant systems.15 While the SUN proteins are highly conserved in eukaryotes, the KASH proteins are less so and it was challenging identifying KASH proteins in plants. Structural hallmarks of KASH proteins include a C-terminal transmembrane domain followed by a short C-terminal domain that resides in the periplasm, contains a highly conserved proline residue and interacts with the SUN domain. This very C-terminal domain is referred to as the KASH domain. The SUN interaction is required for ONM localization of the KASH protein. Zhou et al.16 provided the first characterization of such KASH proteins in plants—the tryptophan-proline-proline (WPP) interacting proteins (WIPs). The WIPs were previously known to be ONM-localized and required for plant-specific Ran GTPase activating protein (RanGAP) anchoring at the NE and NPCs.17,18 At the conference, both Meier and Graumann showed that WIPs and SUN proteins interact, that these interactions are facilitated by the SUN domain and the VVPT domain of WIP1, making this a de facto KASH domain.16 The interaction is not only required to anchor WIP to the NE, but also RanGAP, providing evidence that these SUN-KASH complexes play a role in plant RanGAP anchorage.16 Apart from this novel, plant-specific function, the SUN-WIP complexes are also involved in more conserved mechanisms such as maintaining nuclear shape as a double SUN knock out and a triple WIP knockout lead to change in nuclear shape causing nuclei to become rounder.16 While nucleoskeletal and cytoskeletal associations with these plant SUN-KASH complexes need to be investigated, preliminary evidence presented by Graumann indicate possible AtSUN-NMCP/CRWN linkages similar to SUN-lamin linkages in metazoans (unpublished observation).
In addition, Xiao Zhou (Ohio State University, USA) presented the pain-stakingly identified wifi quintuple mutant, in which all three WIPs and both WPP interacting tail proteins (WITs) are knocked out. Previous research showed that WIP and WIT proteins interact with each other and that these oligomers are required for RanGAP anchorage in undifferentiated root tip tissue. This was shown by loss of RanGAP anchorage at the nuclear periphery in the wifi quintuple mutant.18 Perplexingly the wifi mutant does not have any significant developmental growth defects, providing fresh insight into the mode of action of the RanGAP protein. However outcrossing the wifi mutant reveals that in wifi anthers the male gametophyte does not develop correctly (unpublished observation). Overall, the research from these various groups shows that SUN proteins are key components at the plant NE, similar to their status in non-plant systems, and are involved in numerous protein interaction networks including membrane-intrinsic NE components, nucleoskeletal and NPC components. These interactions likely represent the tip of the iceberg, and the congruence of findings across laboratories and plant models underscore the significance of these advances as widely relevant.
Kentaro Tamura (Kyoto University, Japan) had previously made a significant contribution to the knowledge of the nuclear membrane by identifying and characterizing components of the plant NPC (Fig. 1).19 His research highlighted the similarities and differences of NPC composition between plant and other eukaryotes, supporting the view that most of the core sub-complexes are conserved. However Tamura presented information regarding plant-specific nucleoporin 136 (NUP136), showing by fluorescence recovery after photobleaching (FRAP) that NUP136, in contrast to NUP93, NUP88, GP210 and NUP54, is a mobile nucleoporin, being able to position both at the NPC and in the nucleoplasm.19
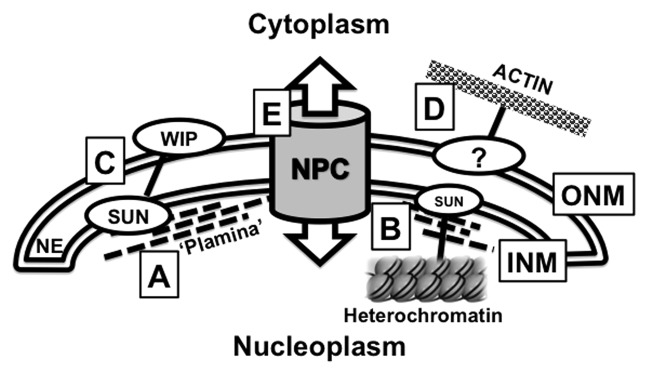
Figure 1. Functional components of the Plant Nuclear Envelope. This meeting highlighted research that focused on the interactions that occur at the plant nuclear envelope (NE). The NE forms a double membrane separating the nucleoplasm and cytoplasm consisting of the Inner Nuclear Membrane (INM) and the Outer Nuclear Membrane (ONM). (A) The NMCP/CRWN proteins are thought to form a proteinaceous “matrix” that controls the shape of the nucleus and is defined in this review as the “Plant Lamina” or “Plamina.” (B) The plamina is hypothesized to associate with SUN proteins that sit within the INM. These SUN proteins also anchor heterochromatic structures such as telomeres and chromocenters to the nuclear periphery. (C) SUN proteins interact with WIPs to form nucleo-cytoplasmic bridges that cross the NE, where the WIP proteins are functional plant KASH proteins. (D) In non-plant systems, nucleus positioning is determined by interactions with the cytoplasmic actin network. However the specific nature of these actin-interacting proteins are currently unknown in plants. (E) The Nuclear Pore Complex (NPC) is an essential conduit for nucleo-cytoplasmic transport. The composition of the plant NPC is similar to equivalent structures in metazoans and yeast yet evidence is emerging for plant specific nucleoporins, such as NUP136. Appropriate references can be found within the main text.
Mutations of plant nucleoporins are known to affect plant processes ranging from development to external stimuli signaling pathways and pathogen interactions.20-23 Geraint Parry (University of Liverpool, UK) explored this topic more closely by presenting three broad classes of nucleoporin mutants that either cause significant developmental phenotypes, have no effect on growth or cause lethality at some stage of the life cycle.24 By making double and higher order mutants, Parry is aiming to create a genetic network of plant nucleoporin function. Parry also found there was little gene expression changes in certain seven-day-old nup mutants when compared with wildtype, but presented the intriguing possibility of a feedback relationship whereby expression of nuclear transport components was upregulated in nup mutants.
Two novel, NE-associated plant proteins were introduced by Marie-Edith Chaboute (IBMP, CNRS, France) as two small (7 kDa) Arabidopsis GCP3 Interacting Proteins, GIP1 and GIP2.25 GCP3 is a component of the γ-tubulin ring complex (γ-TURC), involved in microtubule assembly in plants at the nuclear envelope,26 in yeast spindle pole bodies27 and in the centrosomes of metazoans,28 all corresponding to microtubule organizing centers (MTOCs). Both GCP3 as well as GIP1/2 are localized at the nuclear periphery.25,26 GIP is conserved in all eukaryotes, except S. cerevisiae but the presence of 2 GIP proteins, GIP1 and GIP2 appear limited to plants. Gip1gip2 double mutants demonstrate pleiotropic defects including non-flowering and sterility. At the molecular level, gip1gip2 nuclei are abnormally shaped and their DNA content is significantly increased. GIP1 localizes to the nuclear periphery in interphase nuclei and both GIP1 and SUN1 distributions seem to be correlated.25 Currently the mechanism by which GIP1 contributes to nuclear envelope formation remains unclear, but genetic evidence clearly reveals another possible cytoplasm-nucleoplasm connection, possibly involving SUN domain or associated proteins.
The intense efforts of researchers studying plant NE associated components and processes has already highlighted many similarities and differences to metazoan and yeast systems and will significantly contribute to the field globally. On the whole, the NPC appears to be broadly conserved across eukaryotes yet there is an expectation that plant-specific members will be identified following further proteomic characterization.19 One area where significant differences appear throughout eukaryotes is in the proteins involved in nuclear positioning, a key determinant of cellular function. Whereas SUN domain proteins are present in all systems, the linkage with cytoplasmic partners appears to be plant-specific. WIP proteins take on an equivalent role to metazoan KASH-domain proteins while newly identified SUN-interacting proteins by the Tatout and Evans groups await further characterization. Arguably the most exciting development comes in the identification of the putative plamina, which has been absent from previous characterizations of the plant nucleus. This structure would appear to fulfill the fundamental need for determining the shape of the plant nucleus but the specific biochemical, structurally and functional aspects of this protein matrix are yet to be determined. Given the significant developmental role of the animal lamina, over the next decade it will be fascinating to discover how this plamina contributes to functions of plant cells and its relevance to other eukaryotic systems.
Overall, the research presented at this meeting was of excellent quality and a convincing demonstration of how far and fast this field has moved forward in the last years. Participants expressed great enthusiasm at the resulting recognition that research in plants on the nucleus, nuclear envelope, and cytoplasm has reached a new level. Enabling these breakthroughs was a combination of serendipity to some degree, but also the power of international collaborations, sharing of biological resources, and open exchange of new ideas and unpublished findings to a large degree. As a result, a new working group has been forged, the International Plant Nucleus Consortium. This group aims to take this momentum forward and promote further exchange of ideas and research interactions to shed light on the multifaceted and rapidly advancing biology of the plant nucleus.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/23385
References
- 1.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 2.Fiserova J, Kiseleva E, Goldberg MW. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 2009;59:243–55. doi: 10.1111/j.1365-313X.2009.03865.x. [DOI] [PubMed] [Google Scholar]
- 3.Moreno Díaz De La Espina S, Samaniego R, Yu W, De La Torre C. Intermediate filament proteins with nuclear functions: NuMA, lamin-like proteins and MFP1. Cell Biol Int. 2003;27:233–5. doi: 10.1016/S1065-6995(02)00340-2. [DOI] [PubMed] [Google Scholar]
- 4.Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell. 2007;19:2793–803. doi: 10.1105/tpc.107.053231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruault M, Dubarry M, Taddei A. Re-positioning genes to the nuclear envelope in mammalian cells: impact on transcription. Trends Genet. 2008;24:574–8. doi: 10.1016/j.tig.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 6.van Driel R, Fransz P. Nuclear architecture and genome functioning in plants and animals: what can we learn from both? Exp Cell Res. 2004;296:86–90. doi: 10.1016/j.yexcr.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 7.van Zanten M, Koini MA, Geyer R, Liu Y, Brambilla V, Bartels D, et al. Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc Natl Acad Sci USA. 2011;108:20219–24. doi: 10.1073/pnas.1117726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009;23:928–38. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomita K, Cooper JP. The meiotic chromosomal bouquet: SUN collects flowers. Cell. 2006;125:19–21. doi: 10.1016/j.cell.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 10.Roberts NY, Osman K, Armstrong SJ. Telomere distribution and dynamics in somatic and meiotic nuclei of Arabidopsis thaliana. Cytogenet Genome Res. 2009;124:193–201. doi: 10.1159/000218125. [DOI] [PubMed] [Google Scholar]
- 11.Murphy SP, Bass HW. The maize (Zea mays) desynaptic (dy) mutation defines a pathway for meiotic chromosome segregation, linking nuclear morphology, telomere distribution and synapsis. J Cell Sci. 2012;125:3681–90. doi: 10.1242/jcs.108290. [DOI] [PubMed] [Google Scholar]
- 12.Murphy SP, Simmons CR, Bass HW. Structure and expression of the maize (Zea mays L.) SUN-domain protein gene family: evidence for the existence of two divergent classes of SUN proteins in plants. BMC Plant Biol. 2010;10:269. doi: 10.1186/1471-2229-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friederichs JM, Gardner JM, Smoyer CJ, Whetstine CR, Gogol M, Slaughter BD, et al. Genetic analysis of Mps3 SUN domain mutants in Saccharomyces cerevisiae reveals an interaction with the SUN-Like Protein Slp1. G3 (Bethesda) 2012;2:1703–18. doi: 10.1534/g3.112.004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graumann K, Runions J, Evans DE. Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J. 2010;61:134–44. doi: 10.1111/j.1365-313X.2009.04038.x. [DOI] [PubMed] [Google Scholar]
- 15.Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci. 2009;122:577–86. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X, Graumann K, Evans DE, Meier I. Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J Cell Biol. 2012;196:203–11. doi: 10.1083/jcb.201108098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu XM, Meulia T, Meier I. Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol. 2007;17:1157–63. doi: 10.1016/j.cub.2007.05.076. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Q, Brkljacic J, Meier I. Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell. 2008;20:1639–51. doi: 10.1105/tpc.108.059220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. 2010;22:4084–97. doi: 10.1105/tpc.110.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Coaker G. Nuclear trafficking during plant innate immunity. Mol Plant. 2008;1:411–22. doi: 10.1093/mp/ssn010. [DOI] [PubMed] [Google Scholar]
- 21.Lee Y, Lee HS, Lee JS, Kim SK, Kim SH. Hormone- and light-regulated nucleocytoplasmic transport in plants: current status. J Exp Bot. 2008;59:3229–45. doi: 10.1093/jxb/ern200. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, Hara-Nishimura I. The molecular architecture of the plant nuclear pore complex. J Exp Bot. 2012 doi: 10.1093/jxb/ers258. In press. [DOI] [PubMed] [Google Scholar]
- 23.Parry G. Assessing the function of the plant nuclear pore complex and the search for specificity. J Exp Bot. 2012 doi: 10.1093/jxb/ers289. In press. [DOI] [PubMed] [Google Scholar]
- 24.Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M. The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell. 2006;18:1590–603. doi: 10.1105/tpc.106.041566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janski N, Masoud K, Batzenschlager M, Herzog E, Evrard JL, Houlné G, et al. The GCP3-interacting proteins GIP1 and GIP2 are required for γ-tubulin complex protein localization, spindle integrity, and chromosomal stability. Plant Cell. 2012;24:1171–87. doi: 10.1105/tpc.111.094904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seltzer V, Janski N, Canaday J, Herzog E, Erhardt M, Evrard JL, et al. Arabidopsis GCP2 and GCP3 are part of a soluble γ-tubulin complex and have nuclear envelope targeting domains. Plant J. 2007;52:322–31. doi: 10.1111/j.1365-313X.2007.03240.x. [DOI] [PubMed] [Google Scholar]
- 27.Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–82. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moritz M, Braunfeld MB, Guénebaut V, Heuser J, Agard DA. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–70. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]


