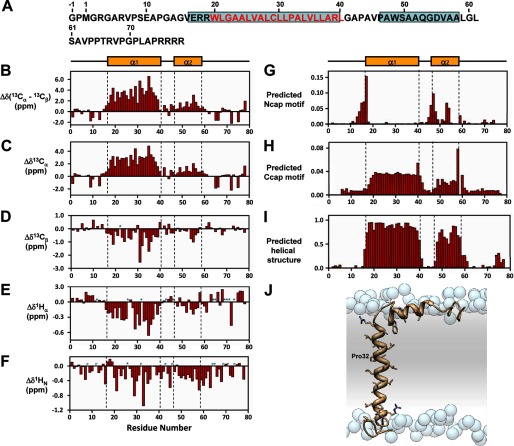
FIGURE 7.
Secondary structure of MMP23-PD. A, amino acid sequence of MMP23-PD where α-helical regions are boxed in cyan and residues in the predicted trans-membrane domain are colored red. B–F, deviations of 13Cα-13Cβ (B), 13Cα(C), 13Cβ (D), 1Hα (E), and 1HN (F) chemical shifts from random coil values (56). Sites for which no values were determined are denoted by a P for proline residues or an asterisk for unassigned. Secondary structure elements derived from CSI values are illustrated at the top and boxed by a dotted line in each figure. Helix α1 extends from residue Glu-17 to Leu-40 and helix α2 from residue Ala-47 to Leu-58. G and H, prediction of N-terminal (G) (Ncap) and C-terminal (H) helical capping (Ccap) motifs using the MICS algorithm. I, helical regions for MMP23-PD determined by MICS (34). MICS is similar to TALOS+ (33), which uses an artificial neural network to predict protein ϕ and ψ backbone torsion angles and protein secondary structure for a given residue along with its two flanking residues using a combination of six kinds of chemical shifts (HN, HA, CA, CB, CO, N) and the amino acid sequence. On the other hand, MICS uses an artificial neural network trained to recognize additional structural features such as N- and C-terminal helical capping motifs and the most common types of β-turns for a given residue (i) within a hexapeptide spanning residues i - 1 to 1 + 4. J, model of MMP23-PD in a lipid membrane. Shown is a model of TMD (α1 helix) and juxtamembrane (α2) helix of MMP23-PD in a lipid membrane. Atoms of the side chain groups of residues in the two helices are highlighted. Phosphate groups of the lipid are represented as blue spheres; lipid choline, glycerol and acyl chains, and water are omitted for clarity.