FIGURE 1.
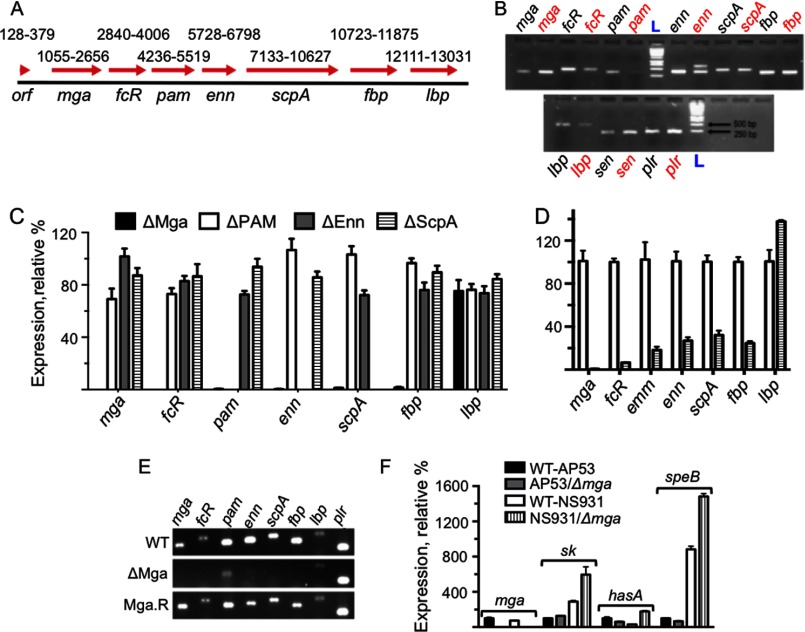
Gene arrangement and expression within the mga regulon region of GAS strains AP53 and NS931. A, total of 13,383 bp from the WT-AP53 genome was sequenced from a genomic clone (black bar; 1–13,383). The red arrows show the genes identified, their directions from 5′ to 3′, and their sequence positions (beginning at the ATG initiation codon to the end of the translation stop codon), arbitrarily starting at position 1 of the assembled genomic clone. The numbering above each arrow shows the range of bp from its translation initiation codon to the translation stop codon. orf is the open reading frame of the most 5′ gene with unknown function; mga is the trans-acting positive regulatory gene for this core group of virulence proteins; fcR, which in the AP53 strain and GAS strain has a stop codon after amino acid 84 from the translation initiation site in an otherwise complete fcR gene, is the antiphagocytic IgG receptor gene; pam refers to the direct Pg-binding gene; downstream of pam is the enn gene, encoding an IgA-binding protein; which is sequentially followed by genes encoding C5a peptidase (scpA), fibronectin-binding protein (fbp), and laminin-binding protein (lbp). Subclones generated by PCR from homologous sequences found in other GAS strains were used to obtain overlapping gene and flanking sequences in order that the genes in the entire genomic clone were identified and positioned. B, PCR amplicons of individual genes in gDNA of WT-AP53 (black) and WT-NS931 (red) gDNA using the PCR primers (supplemental Table 1) for WT-AP53 gene sequences in A. The pam gene is not observed in NS931 cells. C, Q-RT-PCR analysis of the relative transcript levels of the mga core regulon genes in total mRNA isolated from AP53/ΔMga cells (black bars), AP53/ΔPAM (white bars), AP53/ΔEnn (gray bars), and AP53/ΔScpA (striped bars), based on each gene of WT-AP53 arbitrarily set at 100% (not shown in the figure). The WT-AP53 data were first normalized to the gapdh transcript level or that of a second housekeeping gene, mutS, in the individual total mRNA. No significant differences were found whether gapdh or mutS was used for this purpose. D, Q-RT-PCR analysis of the relative transcript levels of the mga regulon genes in total mRNA isolated from WT-NS931 cells (white bars) and NS931/ΔMga cells (striped bars), based on each gene of WT-NS931 arbitrarily set at 100%, as normalized to the gapdh transcript level in the individual total mRNA. The same primers for each gene were used in each WT and isogenic strains. Specific primers for the emm gene were used in this case. E, RT-PCR amplicons for mga, fcR, pam, enn, scpA, fbp, lbp, and plr (gapdh) examined in the isogenic strains, WT-AP53, AP53/ΔMga, and AP53/Mga.R. F, effect of inactivation of the mga gene on critical virulence genes outside of the mga core regulon in WT-AP53 and WT-NS931 cells. The % expression of the specific transcripts in total mRNA is relative to each gene set at 100% in WT-AP53 cells. Primers for individual genes between AP53 and NS931 strains were exact matches to the respective gene sequences. Data are shown for mRNA of sk, the hasA component of the hasABC regulon, and the cysteine protease speB.