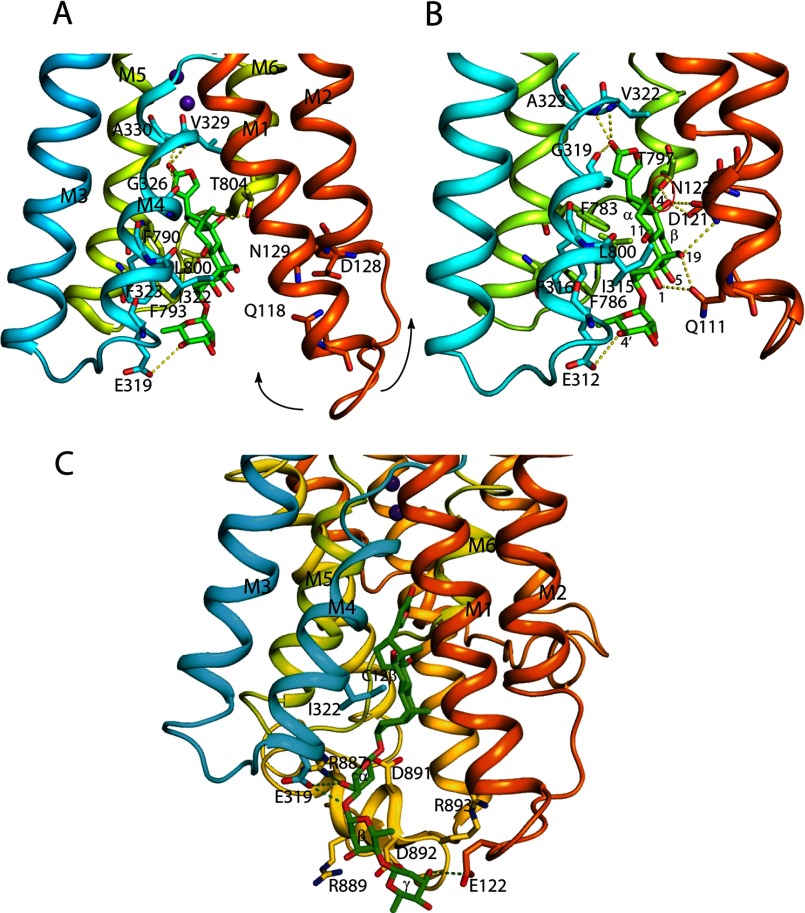
FIGURE 12.
The Na,K-ATPase binding cavity for ouabain. Residues of the Na,K-ATPase α subunit important for coordination and ouabain are shown in sticks. In A, residues important for ouabain interaction in the low affinity ouabain-bound crystal structure are shown together with the two bound K+ ions represented by purple spheres (PDB 3A3Y); in B, the original high affinity ouabain-bound structure is shown (PDB entry 3N23). The arrows in A indicate how M1/M2 moves in the transition to the high affinity state. Residues (shark Na,K-ATPase numbering in A, pig kidney Na,K-ATPase numbering in B) within hydrogen bond distance are connected by yellow dotted lines. The steric clash between the hydroxyl at C14β and Thr-797 (distance 2.2 Å) is circled in red. In A and B, the α-face of the ouabain steroid is stacked with aromatic residues (Ile-322, Phe-323, Phe-790, Phe-793). In C, digoxin (PDB entry 3B0W) is shown docked onto the shark Na,K-ATPase with the steroid core A- and B-rings matching that for ouabain found in the 3A3Y crystal structure. The Na,K-ATPase residues Glu-122, Glu-319, and Arg-887 that are within hydrogen bond distance from hydroxyls of α-, β-, and γ-sugar residues are indicated by dotted lines. In the structures, the β and FXYD subunits are omitted for clarity. The structures were drawn with PyMOL.