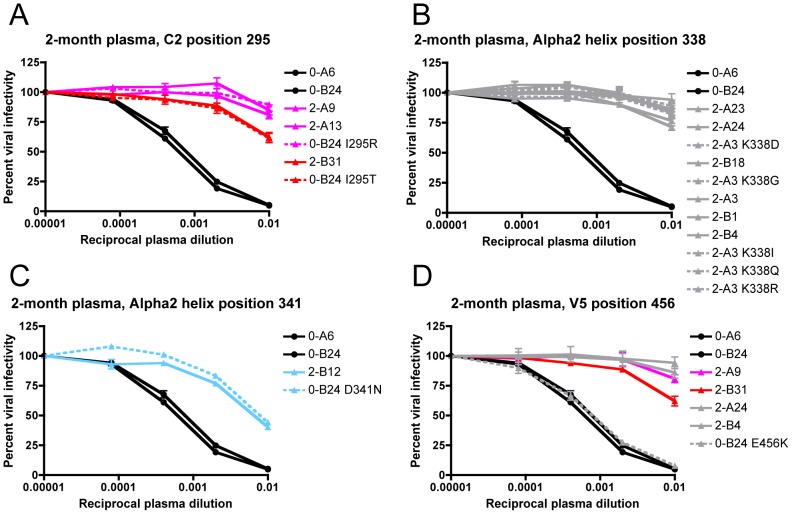
Figure 3. Determination of the earliest R880F nAb escape signatures in Env.
Suspected 2-month nAb escape mutations from four different amino acid positions were introduced by site-directed mutagenesis into the transmitted/founder Env 0-B24 or escape Env 2-A3, which differed from the founder only at the site of mutation introduction. The transmitted/founder and wild-type 2-month Envs (solid lines) along with site-directed mutant Envs (dashed lines) were pseudotyped and assayed with 2-month plasma to determine if substitutions at C2 295 (A), alpha2 helix 338 (B), alpha2 helix 341 (C), or V5 456 (D) (HXB2 residues 293, 337, 340, or 460) could individually confer resistance. Percent viral infectivity, as adjusted against wells containing no test plasma, is depicted on the vertical axis; reciprocal plasma dilutions are plotted along the horizontal axis in a logarithmic fashion. Each curve represents a single Env-plasma combination, and error bars demonstrate the standard error of the mean of two independent experiments using duplicate wells (0-month Envs = circles, 2-month Envs and representative point mutants = triangles). Colored lines (2-A9/2-A13/0-B24 I295R in magenta, 2-B31/0-B24 I295T in red, and 2-B12/0-B24 D341N in cyan) indicate Envs that succumbed to neutralization, in varying combinations, by the isolated R880F mAbs.