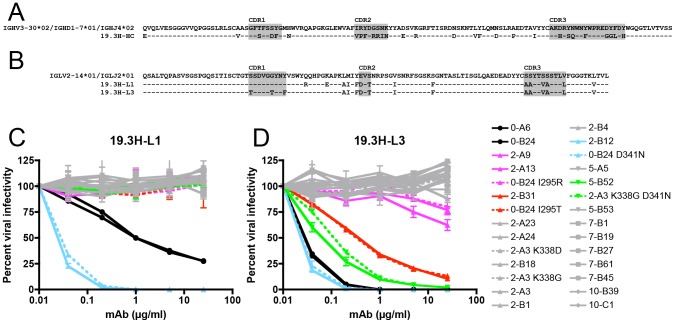
Figure 5. Amino acid alignment of R880F immunoglobulin heavy and light chain variable domains and neutralization by R880F mAbs 19.3H-L1 and 19.3H-L3.
Germline heavy and light chain gene segment utilization was determined by SoDA, a somatic diversification analysis program [32], and amino acid sequences were aligned and examined using Sequencher v5.0 and Geneious v5.0.3 software. Dashes represent conserved positions. Complementarity-determining regions (CDRs) are highlighted in gray. The two R880F mAbs share a common heavy chain, 19.3H-HC (A), which utilizes V3-30*02, D1-7*01, and J4*02 gene families, while the somatically related light chains 19.3H-L1 and 19.3H-L3 (B) employ V2-14*01 and J2*01 gene families and differ from each other at five positions in and just downstream of CDR1. Heavy chain 19.3H-HC, when paired with either 19.3H-L1 (C) or 19.3H-L3 (D), was evaluated for neutralization against pseudotyped R880F wild-type (solid lines) and site-directed mutant Envs (dashed lines). Percent viral infectivity, as adjusted against wells containing no mAb, is depicted on the vertical axis; mAb concentrations (in µg/ml) are plotted along the horizontal axis in a logarithmic fashion. Each curve represents a single Env-mAb combination, and error bars demonstrate the standard error of the mean of two independent experiments using duplicate wells (0-month Envs = circles, 2-month Envs and representative point mutants = triangles, 5-month Envs and a representative point mutant = inverted triangles, 7-month Envs = squares, 10-month Envs = diamonds). Colored lines (2-A9/2-A13/0-B24 I295R in magenta, 2-B31/0-B24 I295T in red, 2-B12/0-B24 D341N in cyan, and 5-B52/2-A3 K338G D341N in green) indicate Envs that succumbed to neutralization, in varying combinations, by the isolated R880F mAbs.