Table 2. Summary of the models presented in this analysis.
| Model |
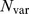
|
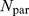
|
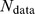
|
Host | Regen. | Abs/CTL/IFN | Other cells |
| Bocharov [2] | 13 | 49 | 19 | Human | yes | Abs/CTL/IFN | Antigen-presenting macrophages, T and B helper cells, B cells, plasma cells, IFN-producing macrophages. |
| Baccam [4] | 6 | 9 | 36 | Human | no | IFN | — |
| Hancioglu [31] | 10 | 28 | 0
|
Human | yes | Abs/CTL/IFN | Antigen-presenting macrophages, plasma cells. |
| Lee [22] | 15 | 48 | 42 | Mouse | yes | Abs/CTL | Dendritic cells, naive and effector CD4 T cells, naive CD8 T cells, naive CD8 T cells, naive and activated B cells, Long- and short-lived plasma cells. T cells, naive and activated B cells, Long- and short-lived plasma cells. |
| Handel [6] | 6 | 8 | 50 | Mouse | yes | Abs/IFN | — |
Miao [23]

|
6 | 8 | 64 | Mouse | yes | Abs/CTL | — |
Saenz [32]

|
8 | 12 | 96 | Horse | no | IFN | — |
Pawelek [37]

|
5 | 11 | 90 | Horse | no | IFN | — |

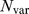 (number of variables),
(number of variables), 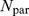 (number of parameters),
(number of parameters), 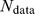 (number of data points), Regen. (whether the model includes cell regeneration).
(number of data points), Regen. (whether the model includes cell regeneration).
 Model was not mathematically fit to data, but did have to conform to some general criteria.
Model was not mathematically fit to data, but did have to conform to some general criteria.
 We consider two different parameter sets for this model (Miao split and Miao full). The differences between these two models are described in Methods.
We consider two different parameter sets for this model (Miao split and Miao full). The differences between these two models are described in Methods.
 The Saenz and Pawelek models were fit to the same data.
The Saenz and Pawelek models were fit to the same data.
