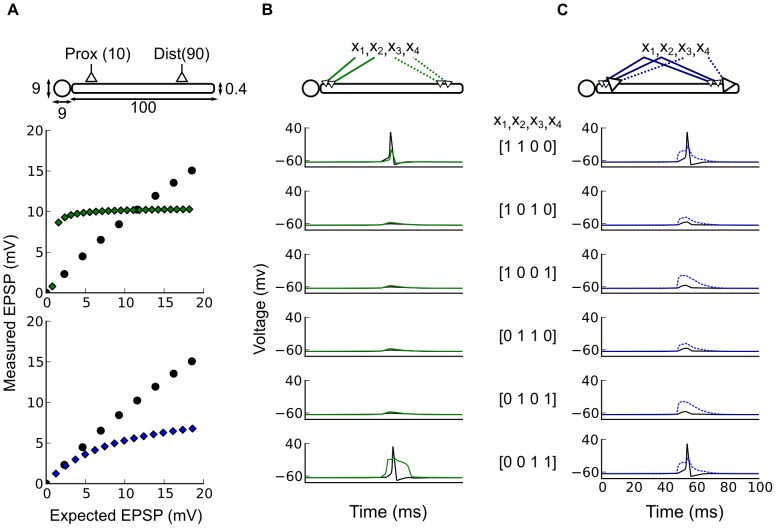
Figure 4. Reduced biophysical models can implement linearly non-separable functions using dendritic saturations or dendritic spikes.
(A) The biophysical model. Upper panel: schematic representation of the biophysical model, where synaptic inputs are clustered (Prox and Dist), and its morphological parameters (input locations, diameters, and dendritic length are in 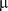 m). Below, expected arithmetic sum versus measured somatic EPSPs: for peri-somatic AMPA stimulation (black dots) producing a linear EPSP integration; for distal NMDA stimulation (green dots) producing a spiking type non-linear summation; for distal AMPA stimulation (in blue) producing a saturation type non-linear summation. (B) Implementation of the feature binding problem using NMDA receptor synapses for the distal dendritic region, illustrating the DNF/local strategy of synaptic placement. Top panel shows how each input makes synaptic contacts in a 10
m). Below, expected arithmetic sum versus measured somatic EPSPs: for peri-somatic AMPA stimulation (black dots) producing a linear EPSP integration; for distal NMDA stimulation (green dots) producing a spiking type non-linear summation; for distal AMPA stimulation (in blue) producing a saturation type non-linear summation. (B) Implementation of the feature binding problem using NMDA receptor synapses for the distal dendritic region, illustrating the DNF/local strategy of synaptic placement. Top panel shows how each input makes synaptic contacts in a 10 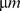 zone either on the peri-somatic or on the distal dendritic region. Below, voltage traces (black∶soma; green∶distal dendrites) in response to the various input patterns. Each voltage trace corresponds to stimulation by a different input vector where an active input variable is a neural ensemble of 100 neurons firing nearly synchronously in a 10 ms window. (C) Implementation of the feature binding problem using only AMPA synapses corresponding to a saturation type non-linear sub-unit and a CNF/global strategy. Top panel shows how each input makes synaptic contacts in a 10
zone either on the peri-somatic or on the distal dendritic region. Below, voltage traces (black∶soma; green∶distal dendrites) in response to the various input patterns. Each voltage trace corresponds to stimulation by a different input vector where an active input variable is a neural ensemble of 100 neurons firing nearly synchronously in a 10 ms window. (C) Implementation of the feature binding problem using only AMPA synapses corresponding to a saturation type non-linear sub-unit and a CNF/global strategy. Top panel shows how each input makes synaptic contacts in a 10 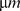 zone either on the peri-somatic or on the distal dendritic region. Below: voltage traces in response to the various input vectors (black∶soma; blue∶distal dendrites).
zone either on the peri-somatic or on the distal dendritic region. Below: voltage traces in response to the various input vectors (black∶soma; blue∶distal dendrites).