Fig. 1.
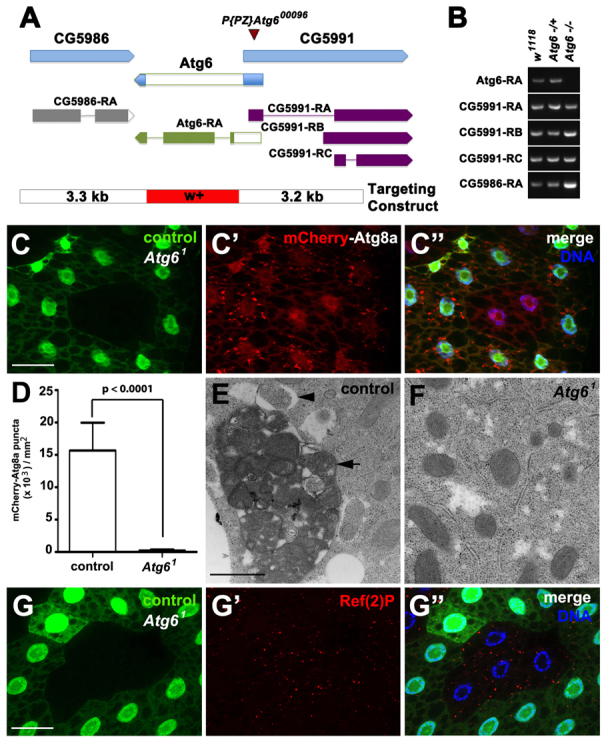
Atg6 is required for autophagy. (A) Atg6 genomic locus and donor targeting construct (not to scale). The donor construct consisted of a w+ mini-gene flanked by 3.5 kb of genomic sequence (blue) from each side of Atg6. Flp recombination target (FRT) sites and I-SceI endonuclease recognition sites were present on each side of flanking sequence, to facilitate double-stranded break and homologous recombination at the target site. The resultant flies contained the w+ minigene (red) in place of the Atg6 ORF (white). (B) RT-PCR of Atg6 and flanking gene transcripts indicates knockdown of Atg6 transcript expression in homozygous Atg61 third instar larvae, and maintenance of flanking gene RNA levels. (C-C″) mCherry-Atg8a puncta reflect starvation-induced autophagosome formation in control (GFP-positive) larval fat body cells, whereas cytoplasmic mCherry-Atg8a localization reflects a defect in autophagy in Atg61 (GFP-negative) fat body cells (n=10). (D) Quantification of mCherry-Atg8a puncta in control and Atg61 mutant fat body cells following starvation. A two-tailed t-test was used for statistical analysis and the P-value relative to control was 8.98×10-8. Error bars represent s.d. (E,F) TEM images reveal abundant autophagosomes (arrowhead) and autolysomes (arrow) in control (E) but not in (F) Atg61 mutant fat body cells following 4 hours of starvation. (G-G″) Ref(2)P accumulated in Atg61 (GFP negative) fat body cells, indicative of defective autophagy, compared with control cells (GFP positive) (n=13). Scale bars: 50 μm in C-C″,G-G″; 500 nm in E,F.
