Abstract
Background
Mice harbouring gene mutations that cause phenotypic abnormalities during organogenesis are invaluable tools for linking gene function to normal development and human disorders. To generate mouse models harbouring novel alleles that are involved in organogenesis we conducted a phenotype-driven, genome-wide mutagenesis screen in mice using the mutagen N-ethyl-N-nitrosourea (ENU).
Methodology/Principal Findings
ENU was injected into male C57BL/6 mice and the mutations transmitted through the germ-line. ENU-induced mutations were bred to homozygosity and G3 embryos screened at embryonic day (E) 13.5 and E18.5 for abnormalities in limb and craniofacial structures, skin, blood, vasculature, lungs, gut, kidneys, ureters and gonads. From 52 pedigrees screened 15 were detected with anomalies in one or more of the structures/organs screened. Using single nucleotide polymorphism (SNP)-based linkage analysis in conjunction with candidate gene or next-generation sequencing (NGS) we identified novel recessive alleles for Fras1, Ift140 and Lig1.
Conclusions/Significance
In this study we have generated mouse models in which the anomalies closely mimic those seen in human disorders. The association between novel mutant alleles and phenotypes will lead to a better understanding of gene function in normal development and establish how their dysfunction causes human anomalies and disease.
Introduction
Genetic approaches in the mouse have been instrumental in aiding our understanding of the development, disease and congenital anomalies of the major organ systems. Most of our current knowledge has arisen through the use of “reverse genetic” or gene-driven approaches in which the gene of interest is mutated and the subsequent phenotype analysed. Although reverse genetics has provided us with a wealth of knowledge it has some limitations. It requires prior information about the function of the gene to be altered, is labour intensive, may result in an unexpected phenotype or no phenotype at all, and can only produce one mouse line at a time [1].
To circumvent some of these limitations, along with the recent advances in sequencing technologies, phenotype-driven (forward genetics) large-scale screens have come back in vogue using the chemical mutagen N-ethyl-N-nitrosourea (ENU). ENU is efficient and potent, inducing ∼1 mutation per 1–2.7 megabase of the genome. It creates point mutations, in coding or splice site regions, in a relatively unbiased fashion. These mutations better reflect the types of mutations responsible for human disease. ENU can induce different point mutations within the same gene, thereby creating an allelic series, each able to reveal a different aspect of the protein’s function which would not be revealed by a null allele. ENU injected into male mice introduces mutations into the genome of all cells including the spermatogonial stem cells allowing the mutations to be transmitted through the germ-line. Thousands of genes can be mutated in an efficient manner and mice screened for phenotypes of interest. Once inheritance of a phenotype is established the causal gene can be identified using linkage analysis and sequencing approaches [2], [3], .
Until recently, the bottleneck of ENU mutagenesis has been the identification of the causal gene. However, recent technological advancements in mapping and sequencing have made identification of the mutant gene more rapid. Typically, gene identification has previously been reliant on outcrossing the affected male to a different inbred strain. The progeny are then backcrossed or intercrossed. The chromosomal region containing the causal gene can then be identified by linkage analysis. Linkage analysis technologies are now available in high throughput formats using mouse strain specific single nucleotide polymorphisms (SNPs). Once linkage to a chromosomal region is identified candidate gene sequencing can be carried out. If candidates are not evident then recent advancements in next-generation sequencing technologies have made sequencing the exons of hundreds of genes within a chromosomal region feasible [8], [9], [10]. Alternatively to chromosomal linkage analysis whole exome sequencing may be used to identify the causal gene and point mutation [7].
Many ENU screening strategies have been employed. These include region-based screens to identify mutations in a defined region of the genome [11], [12], [13], [14], [15], [16], [17], sensitized screens [18], [19], [20], [21] and genome-wide screens [22], [23], [24], [25], [26], [27]. Many genome-wide, phenotype based screens have been performed to identify both dominant and recessive ENU-induced mutations. These screens have been extremely successful in identifying a range of novel alleles important for embryonic development and disease. Given that many recessive mutations are lethal it is necessary to perform these screens embryonically. Genome-wide embryonic recessive screens have focussed on specific developmental time points and specific phenotypes of interest [28], [29], [30], [31], [32], [33], [34].
In the present study we performed an ENU mutagenesis screen in embryonic mice to identify recessive mutations in genes involved in organogenesis. We brought together a team of developmental biologists (ENU Organogenesis Consortium), each with expertise in the development of a specific mouse organ system. We screened G3 embryos at embryonic day (E) 13.5 and E18.5 with a focus on identifying phenotypic abnormalities in limb and craniofacial structures, skin, blood, vasculature, lungs, gut, kidneys, ureters and gonads. A hierarchical and systematic strategy was developed in order to screen each of these structures/organs in every G3 embryo. We identified 15 mutant mouse lines exhibiting abnormalities in one or more of the structures/organs screened for and identified novel mutant alleles for Fras1, Ift140 and Lig1.
Results
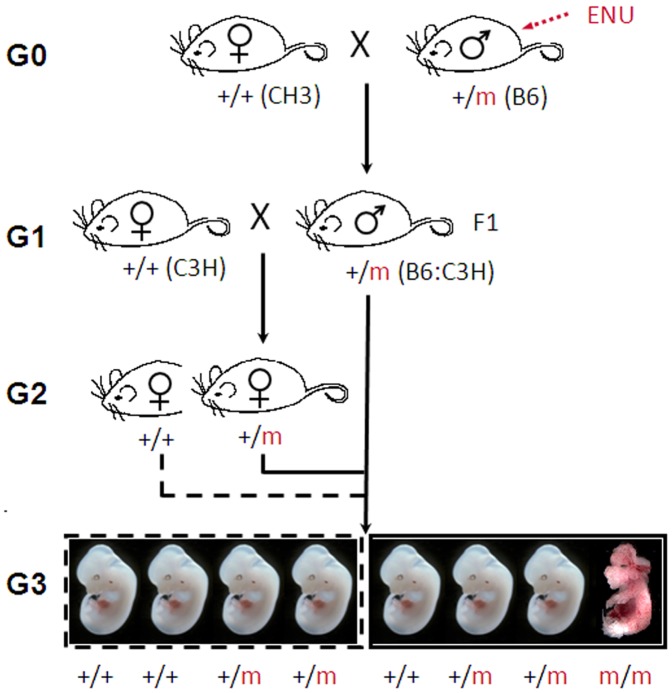
To screen for recessive mutations a standard three generation breeding protocol was used (Figure 1). Of the 52 pedigrees screened 25 were screened at E13.5 only, 14 were screened at E18.5 only and 13 were screened at both time points. We identified 15 pedigrees with a reproducible phenotype in multiple litters (Table 1). Embryos with a range of anomalies were identified including, structural anomalies (craniofacial, skeletal and limb defects, exencephaly, spina bifida and curled tail) and organ anomalies in the kidney, ureter, lung, skin, gonad, eye and blood. Line 12BCC-22 carried two mutations that resulted in the segregation of two pedigrees with distinct phenotypes (anaemia and spina bifida) (Table 1).
Figure 1. Breeding strategy for the generation of G3 embryos.
C57BL/6J male mice were injected with ENU and bred with females of the inbred mapping strain, C3H/HeH (C3H). +/+ = wildtype, m = mutation, G = generation.
Table 1. Summary of ENU-induced mouse mutants.
| Line | Time-point | Phenotype | Affected/Total no.embryos screened* | Affected/Total no. of embryos in affected litters | Gene ID/mapping |
| bfb # | 18.5 | skeletal (craniofacial/polydactyly) | 16/149(11%) | 9/46(20%) | Fras1 |
| skin | 5/37 (3.5%) | ||||
| kidney | 2/21 (9.5%) | ||||
| 11BC-11 | 18.5 | Skeletal (syndactyly, craniofacial) | 5/116 (4%) | 5/20 (25%) | Unknown |
| snoopy# | 13.5 | craniofacial | 8/134 (6%) | 8/32 (25%) | Unknown |
| kanyon# | 13.5 | craniofacial, neural tube, exencephaly | 12/52 (23%) | 12/26 (46%) | Chr 7 |
| cauli # | 13.5 | craniofacial, exencephaly, spina bifida, polydactyly, limb anomalies | 15/73 (20.5%) | 15/54 (28%) | Ift140 |
| 12BC-19 | 18.5 | Kidney, ureter | 12/208b (6%) | 12/64 (18.8%)b | unknown |
| 12BC-20 | 18.5 | Kidney, ureter | 11/198b (6%) | 11/42 (26%)b | unknown |
| 12BCC-13 | 18.5 | lung | 7/176 (4%) | 7/21 (33%) | unknowna |
| 12BCC-16 | 18.5 | lung | 3/27 (11%) | 3/8 (37.5%)$ | unknowna |
| 12BCC-20 | 13.5 | gonad | 7/280b (2.5%) | 7/36 (19%)b | unknowna |
| 12BCC22a# | 13.5 | blood | 11/136 (8%) | 11/29 (38%) | Lig 1 |
| 12BCC-22b | 13.5 | spina bifida, curled tail | 23/136 (17%) | 23/60 (38%) | unknown |
| 12BCC-24 | 18.5 | curled tail | 4/24 (17%) | 3/15 (20%) | unknown |
| fused digit | 1/15 (6.7%) | ||||
| 14BC-2 | 18.5 | eye pigmentation | 3/31 (10%) | 3/15 (20%) | Unknown |
| 14BC-7 | 18.5 | Kidney, ureter | 7/305b (2%) | 7/43 (16%)b | Unknown |
Allele names and symbols have been registered with MGI. Allele ID for Ift140cauli is MGI: 4458412. Mouse strains have been deposited into the Australian Phenome Bank repository (http://pb.apf.edu.au/phenbank).
Number of G3 embryos screened in litters with ≥6 embryos/litter.
Affected embryos in 3 individual litters. Litter sizes were <6 embryos for this pedigree.
The phenotype was not inherited by the 2nd generation males.
Total numbers include male and female. Testis anomalies are only seen in males and kidney anomalies are predominately seen in males. Therefore a greater number of total embryos were screened.
A Novel Fras1 Allele Displaying Craniofacial, Skeletal, Skin and Urinary Tract Anomalies
Line 11BC-5 was originally identified as a strain that exhibited isolated cleft secondary palate at E18.5. Additional screening revealed a range of phenotypes including blood filled blisters on the head and feet at E13.5 and preaxial polydactyly, open eye lids (Figure 2A–D) and renal agenesis (data not shown) at E18.5. Based on the phenotype observed in E13.5 embryos, this strain was named blood filled blisters (bfb). This collection of phenotypic features is characteristic of the blebs mutants that arise following mutation of Fras1, Frem 1, 2 and Grip1 [35]. Targeted SNP analysis using markers polymorphic between C57BL/6 and C3H were used to investigate the possibility of linkage of the bfb phenotype to one of these four blebs genes using a cohort of 9 phenotypically mutant embryos. Markers flanking Frem1, Frem2 and Grip1 exhibited mixed genotypes (Figure 3A). In contrast, markers flanking Fras1 were homozygous for the C57BL/6 allele in 9/9 samples, indicating clear linkage of the bfb phenotype to Fras1. Sequencing of all 75 exons and flanking sequences from genomic DNA of a single mutant identified a 10762T>C substitution (open reading frame of NCBI RefSeq transcript NM_175473.3) causing a Ser3588Pro mutation in FRAS1. PolyPhen-2 analysis predicts that this change is “probably damaging” and SIFT predicts that it is “not tolerated”.
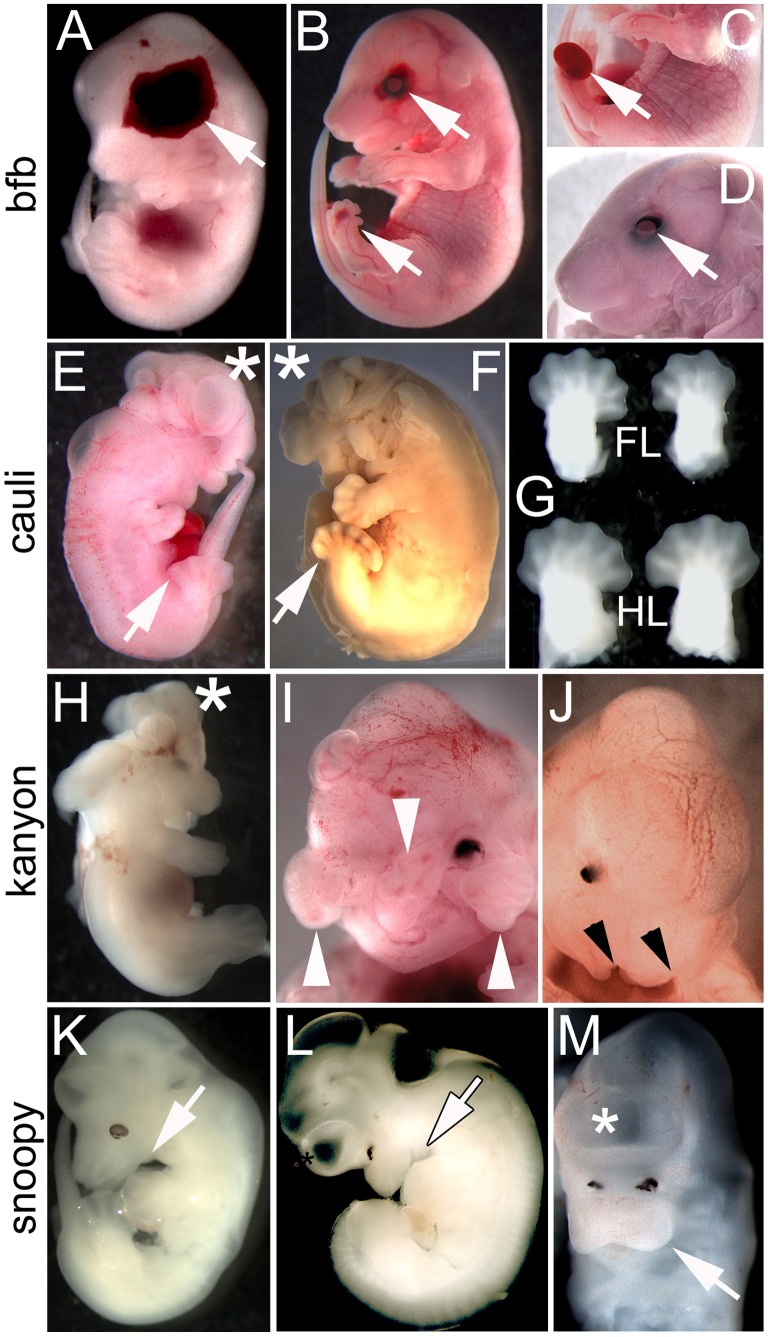
Figure 2. Craniofacial and skeletal mutants.
(A–D) 11BC-5 (blood filled blisters; bfb), (E–G) 11BC-3 (cauli), (H–J) 12WT-49 (kanyon), (K–M) 12WT-9 (snoopy). bfb mutant at E13.5 (A) and E18.5 (B–D) exhibiting characteristic haemorrhagic blisters over the eye, side of the head and feet (arrows). The foot blisters can be discrete or distended as in (C) but are typically associated with digit malformations including polydactyly. The blisters over the eye are commonly associated with open eyelids (D). Cauli embryos at (E) E13.5 and (F) E16.5 present with exencephaly (asterisk) and polydactyly (arrow). (G) Fore- (FL) and hindlimbs (HL) of an E13.5 cauli embryo illustrating the variable autopod phenotype in the forelimbs. Kanyon embryos (H-J) frequently present with exencephaly (H, asterisk) and midfacial clefts. Clefts may result from a defect of frontonasal process development such that the maxillary and frontonasal processes (arrowheads) completely fail to fuse (I) or may present as bilateral cleft lip and palate (J) in mild cases. Regardless of the severity of the facial cleft, the eyes never develop normally (I, J). Snoopy embryos (K-M) present with forebrain malformation, poor eye development and mandibular hypoplasia/agnathia (arrow). (L, M) The forebrain often fails to divide into two vesicles (asterisk) and is associated with various degrees of hypotelorism (M).
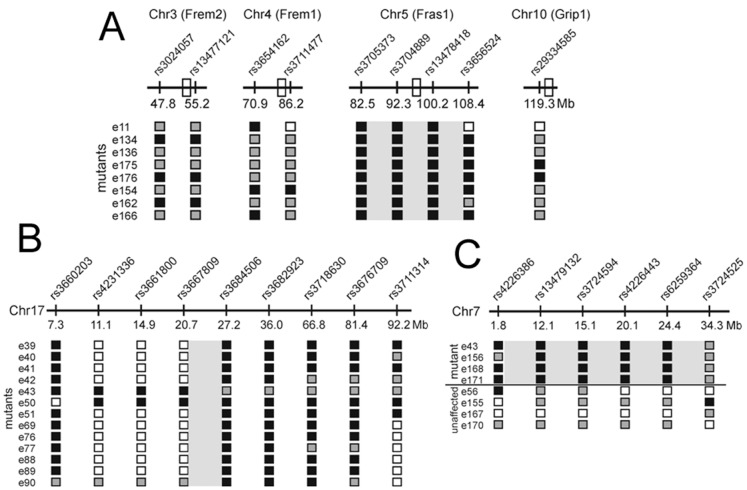
Figure 3. Linkage Analysis.
(A) Based on the phenotype, a targeted approach was used to establish linkage to Fras1 in the bfb strain. Polymorphic markers flanking the four previously characterised blebs genes (open boxes) were used to identify a candidate for sequencing. Strong linkage was observed with the Fras1 gene (grey box). (B) A genome-wide scan established linkage to chromosome 17 in the cauli strain. Chromosome 17-specific markers were then used to refine linkage to a 7 Mb region between rs3667809 and rs3684506 (grey boxed region) containing 218 genes. No additional informative markers were available to further refine the linkage region. (C) A genome-wide scan established linkage to chromosome 7 in the 12BCC-22a line. Subsequent analysis using chromosome 7-specific markers refined linkage to a 32 Mb region containing approximately 700 genes. Numbers to the left in each diagram denote individual mutant embryos except in (C) where both mutant and unaffected embryos are shown. Black boxes denote homozygous C57BL/6 alleles, grey boxes denote C57BL6/C3H heterozygosity, white boxes denote untested markers.
An Ift140 Mutation Resulting in a Complex Ciliopathy
Line 11BC-3 was screened at E13.5 only and exhibits a very consistent phenotype involving exencephaly, anophthalmia, craniofacial malformation, hindlimb polydactyly and forelimb poly- and oligodactyly (Figure 2E–G). The hindlimbs were almost always bilaterally polydactylous while the forelimbs exhibited roughly equal proportions of oligodactyly, normodactyly and polydactyly which was uni- or bilateral (Figure 2G). In reference to the exencephalic morphology, this strain was named cauliflower (cauli). A significant proportion of E13.5 cauli embryos were in poor condition and the incidence of embryonic death increased rapidly with gestational age such that from 22 E17.5–18.5 litters only 7/137 (5.1%) cauli embryos have been recovered. The poor condition of many mid-gestation embryos made investigation of laterality defects impossible but a number of younger embryos have incidentally been observed to have heart looping defects (not shown). The impact of these early defects on later cardiac morphology is yet to be investigated. Conventional SNP-based linkage analysis identified a 7 Mb interval on chromosome 17 containing 218 genes (Figure 3 B). Given that the cauli mutant phenotype strongly resembled the phenotype of Ift144 mutants [36], [37] a strong candidate gene within this interval was, Ift140, a component of the intraflagellar transport complex A (IFT-A) that regulates retrograde protein transport in ciliated cells [38], [39]. A 2564T>A substitution (open reading frame of NCBI RefSeq transcript NM_134126.3) resulting in an Ile855Lys mutation was identified in IFT140. PolyPhen-2 indicates that this change is “possibly damaging” while SIFT analysis predicts that it is “not tolerated”.
Kanyon, a Model of Frontonasal Dysplasia
The 12WT-49 strain phenotype was identified at E13.5 and presented with defects in the frontonasal process, resulting in a mild to severe mid-facial cleft, exencephaly and neural tube defects (Figure 2 H–J). In severe cases the mid- and upper face was completely cleaved and this strain was therefore named kanyon. This phenotype is similar to a group of conditions known as frontonasal dysplasias (OMIM 136760) in which there are deficits in production of the mid-facial skeleton. Interestingly, the facial clefting and exencephaly while commonly observed together could also occur independently. There were no other gross phenotypes observable. Conventional SNP based mapping identified an approximately 67 Mb region on chromosome 7 between marker rs3713432 and rs4226997 containing approximately 1300 genes.
A Mutant Displaying a Link between Holoprosencephaly and Mandibular Outgrowth
The 12WT-9 strain was identified at E13.5 and presented with forebrain anomalies, mandibular hypoplasia/aplasia, poorly developed eyes and hypotelorism (Figure 2 K–M). Given the combination of small eyes and lower jaw, this strain was named snoopy, in reference to its resemblance to the cartoon character of the same name. A small proportion of affected embryos presented with complete failure of forebrain vesicle separation and cyclopia. This suggests that the forebrain anomalies and hypotelorism seen in snoopy are part of a holoprosencephaly spectrum. The aetiology of the failed mandibular outgrowth is unclear and could be secondary to cranial neural tube defects. Mandibular hypoplasia/agnathia is associated with human holoprosencepahly in the condition agnathia-otocephaly complex (OMIM 202650). A whole exome sequencing approach is being used to identify the causative mutation in this strain.
An Isoleucine to Phenylalanine Substitution in DNA Ligase 1 (Lig1) Results in Anaemia
During early mouse development, embryonic blood is first produced in the blood islands of the yolk sac (E8.5–E10.5), then later, during organogenesis (E11.5– E18.5), blood production switches to the fetal liver in order to meet the increasing needs of the circulatory and immune systems. At E13.5 haematopoietic cells constitute at least 50% of the fetal liver and most of these are haemoglobinised erythroid progenitors. Mutations that completely block erythropoiesis, such as Gata-1 or Fog-1 null mutations cause visible anaemia in the liver and circulation by E12.5 and are lethal between E12.5– E15.5 [40], [41].
Based on these criteria we screened embryos for pale livers and identified E13.5 G3 embryos with visible anaemia in the liver in line 12BCC-22a (Figure 4A). Giemsa staining of peripheral blood verified that the embryos lacked mature enucleated red blood cells (RBC) (Figure 4 B, C). The gene responsible for this blood defect was localised to a 32.6 Mb region on the proximal end of chromosome 7 (rs4226386–rs3724525) (Figure 3 C). This region contained approximately 700 genes. Sequencing of all exons in this region using NGS identified a 1798A>T substitution in DNA ligase 1 (Lig1) (open reading frame of NCBI RefSeq transcript NM_001199310.1) resulting in an Ile600Phe mutation. The structure of Human LIG1 has been solved and we used this to predict that Ile 600 is on the surface that contacts DNA during DNA binding and repair [42]. This mutation alters the charge of this amino acid in the DNA binding and ATP dependant DNA ligase domain of LIG1. This may affect the ability of LIG1 to catalyse the joining of single strand DNA breaks during DNA replication, repair and recombination.
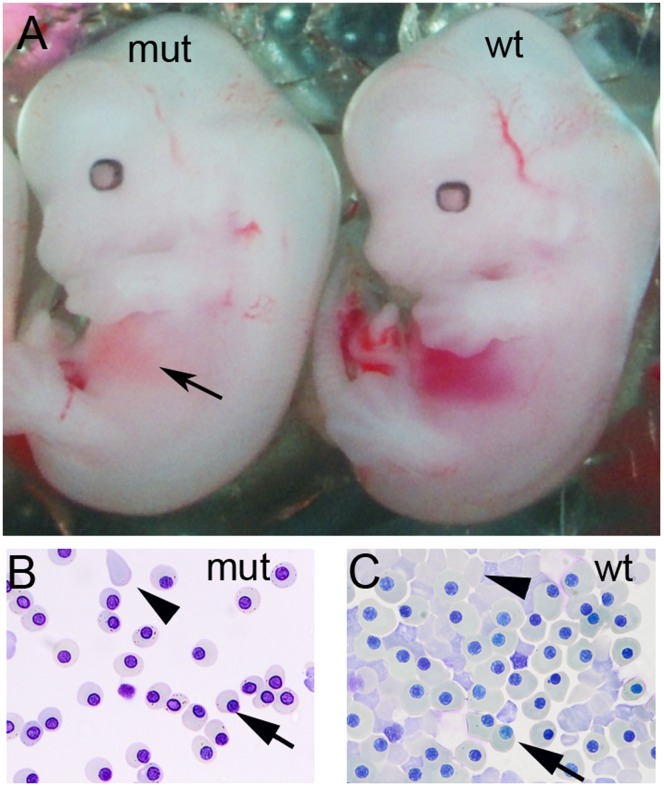
Figure 4. Anaemia in Lig1 mutants.
(A) A Lig1 mutant (mut) with pale orange liver (arrow) compared to the red haemoglobin rich liver of a wildtype (wt) littermate. (B, C) Analysis of peripheral blood in E13.5 embryos by cytospin and Giemsa staining (x40). (B) Lig1 mutant (mut) peripheral blood shows a high proportion of nucleated erythroblasts (arrow) to enucleated erythroblasts (arrowhead) compared with (C) a wild type (wt) littermate.
Line 12BCC-20 Displays Testis Cord Abnormalities
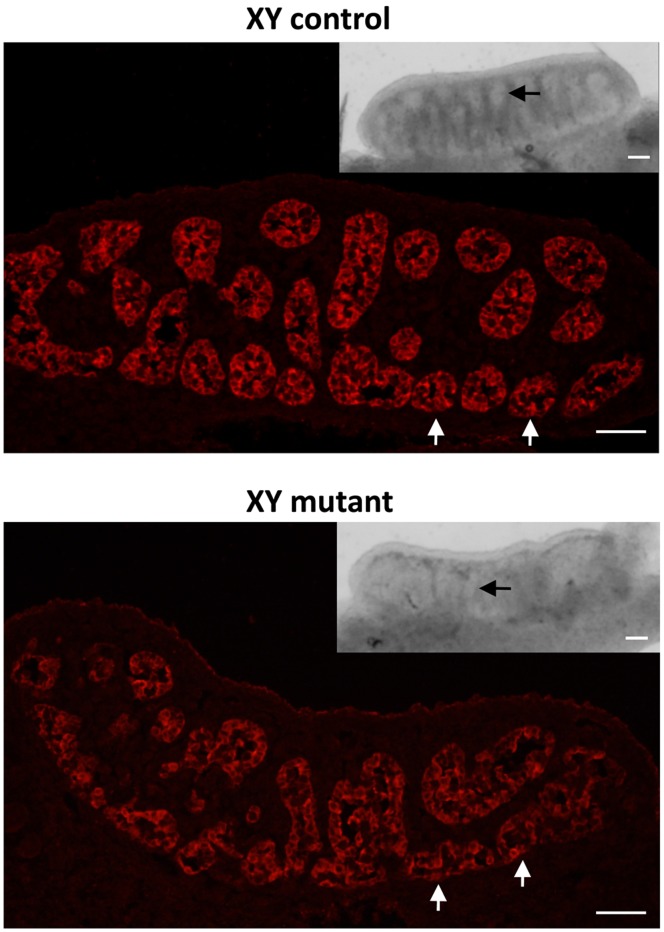
At E11.5, in the mouse, the bipotenital gonad commits to being either male (testis) or female (ovary) [43]. By E13.5, testes can be easily distinguished from ovaries by the presence of testis cords and their bigger size. In this screen we were interested in identifying mutants in which this decision had been disrupted resulting in an ovotestis phenotype. In addition, we were interested in mutations that may disrupt testis size, shape and testis cord formation. In line 12BCC-20, male G3 embryos with smaller testes were identified at E13.5 (7/36 embryos from affected litters (19.4%)) (Figure 5, insets). Moreover, while testis cords were clearly visible in unaffected gonads by gross examination, in affected gonads they were barely detectable (Figure 5, arrows in insets). Immunofluorescence staining for the Sertoli cell marker, AMH, revealed that in affected embryos the testis cords were less were less clearly defined when compared to unaffected embryos (arrows in Figure 5). The embryos with testis cord abnormalities were genotyped for chromosomal sex by PCR. All embryos had an XY karyotype; thus they did not display XX gonadal sex reversal. Unfortunately, the gonad anomalies were not detected in the second generation males possibly due to a weakly penetrant phenotype and the reduced contribution of the C57BL/6 mouse strain. C57BL/6 mice are exquisitely sensitive to testis development defects due to a higher gonadal expression of a female transcriptome relative to other mouse strains [44].
Figure 5. Testis cord defects in line 12BCC-20 at E13.5.
Immunofluorescence staining for the Sertoli cell marker AMH (red, cytoplasm) on testis sections from unaffected (XY control) and affected (XY mutant) embryos. In affected embryos, testis cords are less organized when compared to unaffected embryos. Insets: Gross-examination of testes from unaffected and affected embryos. In affected embryos, testes are smaller and testis cords are barely detectable. Arrows indicate testis cords. All scale bars are 50 µm.
Lines 12BC-19, 12BC-20 and 14BC-7 Display Kidney and Ureter Anomalies
Congenital anomalies of the kidney and urinary tract (CAKUT) commonly arise due to abnormal ureter development as a result of ectopic or lack of budding of the ureteric bud (presumptive ureter), ectopic ureter insertion into the bladder, abnormal smooth muscle cell or urothelium development and obstruction [45]. These defects, which are more commonly found in baby boys [46], [47], can lead to duplicated ureters, multiple kidneys, renal agenesis, hypoplasia and vesicoureteral reflux (backflow of urine into the kidney) causing ureter and renal pelvis dilation and hydronephrosis.
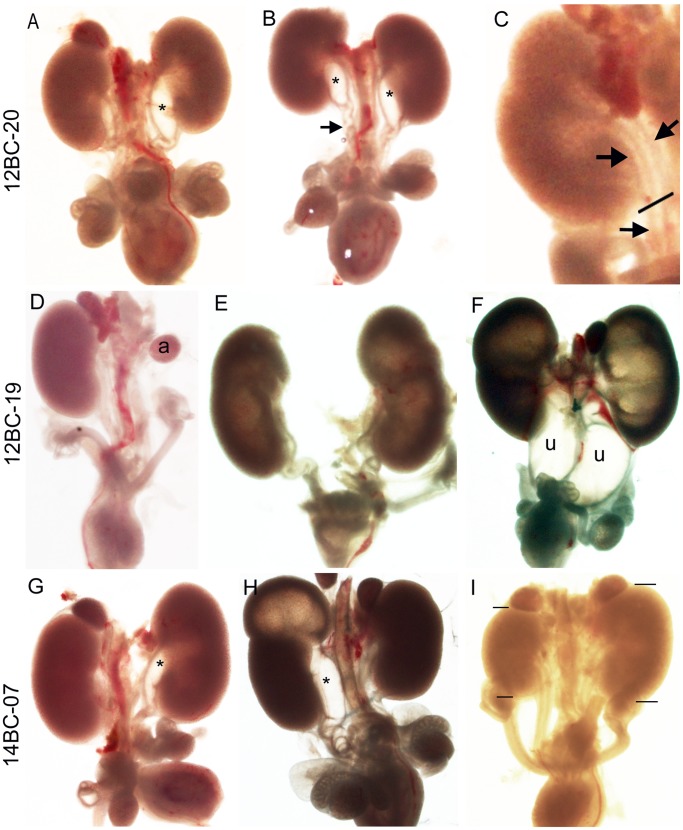
Screening for these various kidney and ureter anomalies identified three affected lines, 12BC-20, 12BC-19, 14BC-7. In all lines we found a higher incidence of renal anomalies in males (82–67%) compared to females (18–33%). Kidneys from males in line 12BC-20 predominantly displayed dilation of the renal pelvis and proximal hydroureter either unilaterally (55%, 6/11 affected embryos) or bilaterally (36%, 4/11 affected embryos) (Figure 6 A–C). Several males had kinked ureters or severely twisted ureters distal to the dilation (36%, 4/11 affected embryos) (Figure 6 A and B). Females displayed either bifurcated ureters (9%, 1/11 affected embryos) (Figure 6 C) or two ureters with one entering the kidney and the other being blind-ended (9%, 1/11 affected embryos). In line 12BC-19 the majority of the affected embryos displayed dilation of the renal pelvis with or without hydroureter and hydronephrosis (58%, 7/12 affected embryos), 17% (2/12 affected embryos) displayed ureter duplication and 25% (3/12 affected embryos) displayed renal agenesis (Figure 6D–F). In line 14BC-7, 43% of the affected embryos displayed dilation of the renal pelvis (3/7 affected embryos), 43% displayed an alteration in kidney size or position which is unique to this pedigree (3/7 affected embryos), 7% displayed ureter duplication with hydroureter/hydronephrosis and 7% displayed unilateral renal agenesis (1/7 affected embryos) (Figure 6G–I). In all three lines the embryos did not exhibit any gross non-renal anomalies.
Figure 6. Urinary tract anomalies displayed by lines 12BC-19, 12BC-20 and 14BC-07.
12BC-20 (A–C) (A) Unilateral and (B) bilateral dilation of the renal pelvis (*). Kinks (A) and twisting (B) of the ureter (arrow) distal to the dilation. (C) Ureter bifurcation at the site marked with a line. Single ureter (arrow) below and bifurcated ureter (arrows) above the line. 12BC-19 (D–F) (D) Unilateral renal agenesis. (E) Bilateral hydronephrosis. (F) Bilateral hydroureter and hydronephrosis. 14BC-07 (G–I) (G) Dilated renal pelvis (*). (H) Duplicated ureter with hydroureter in one ureter leading to hydronephrosis in upper pole. (I) Unilateral renal hypoplasia (lines marking top and bottom of kidneys). a = adrenal, u = ureter.
Mutants with Lung Anomalies
Our primary focus for the lung screen was to identify mutants at E18.5 with thickened lung mesenchyme. The rationale being that during the late stages of fetal life there is a rapid increase in distal airspaces in the lung and a thinning of the mesenchyme between airspaces. These structural changes greatly enhance the gas exchange potential of the lung in preparation for birth when the lung must take on the role of gas exchange for the first time [48], [49]. If the lung mesenchyme fails to thin, it impairs gas exchange after birth and can be fatal in the absence of respiratory support [50].
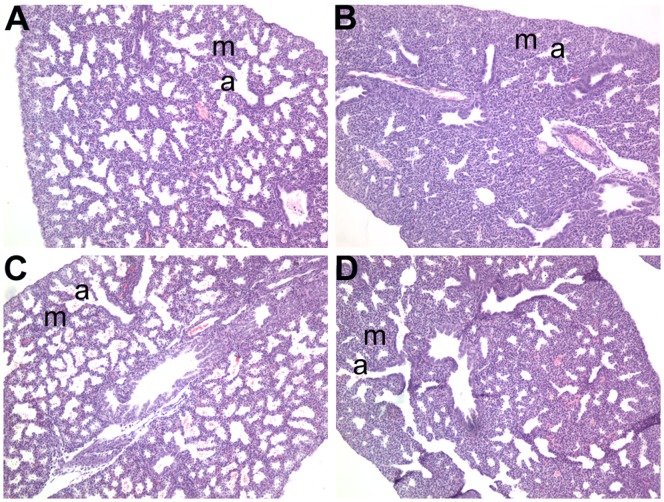
To identify lungs with thick lung mesenchyme a histological screen was undertaken at E18.5 (just prior to birth). We identified two lines 12BCC-013 (7/21 embryos from affected litters, 33%) and 12BCC-016 (3/8 embryos from affected litters, 37.5%) with thick, hypercellular lung mesenchyme (Table 1, Figure 7). These mouse models offered the potential to increase our understanding of the molecular mechanisms that regulate thinning of the lung mesenchyme prior to birth. Unfortunately, the thick hypercellular phenotype was not detected in the second generation mice when searching for carrier males in line 12BCC-013. This may be due to a weakly penetrant lung phenotype or the introduction of a modifier allele that may block the phenotype. Line 12BCC-016 had to be abandoned as the G1 male mouse failed to produce further progeny.
Figure 7. Lung anomalies displayed by lines 12BCC-013 and 12BCC-016.
Lung histology in line 12BCC-013 (A, B) and 12BCC-016 (C, D). Affected lungs in lines 12BCC-013 (B) and 12BCC-016 (D) both displayed hypercellular, thickened lung mesenchyme (m) which can be clearly seen by the increased pink/purple staining of cells and their nuclei, between the small airways (a) in B and D compared to that in the lungs of unaffected littermate embryos (A and C), respectively.
Discussion
The genetic insults responsible for many human congenital defects are yet to be discovered. Generating mouse models of congenital anomalies provides a valuable resource for understanding the aetiology and molecular mechanisms contributing to such congenital defects. Using ENU mutagenesis to generate such mouse models enables novel alleles to be associated with these conditions as well as providing valuable insights into the control of normal development. Our phenotype-driven, genome-wide ENU mutagenesis screen was designed to identify embryonic anomalies across multiple organs and structures within the same set of embryos. In addition, we successfully established a hierarchical and systematic protocol that could be used for future high throughput multi-organ screens. We generated ENU-induced mouse models that displayed craniofacial, skeletal, kidney, ureter, lung, skin, haematological and gonadal anomalies and have identified several novel alleles associated with some of these anomalies.
The most common anomalies identified in this screen involved structural defects (craniofacial, skeletal, limb and neural tube defects). Craniofacial anomalies comprise one third of all human birth defects and have a prevalence of 1% of live births worldwide [51]. Several pedigrees in this study displayed craniofacial anomalies. In particular, the kanyon and snoopy mutants exhibited phenotypes that mimic those seen in the human conditions, frontonasal dysplasias (OMIM 136760) and agnathia-otocephaly complex (OMIM 202650), respectively. Further characterisation of the kanyon and snoopy strains and the genetic mutation underpinning them will provide valuable information on the aetiology of similar conditions in humans.
Several mutants with defects in limb/digit formation were identified in this screen. Of particular interest was the cauli mutant which predominantly displayed exencephaly and polydactyly and is highly reminiscent of the phenotype seen in a number of sonic hedgehog (Shh) signalling pathway mutants [52], [53], [54]. The similarity of the cauli phenotype to existing Shh pathway mutant models provided very strong support for selecting Ift140 as a candidate for sequencing. IFT140 is a core component of the retrograde intraflagella transport mechanism and is therefore a key component of the signalling nexus operating within the primary cilium [38], [39]. Further studies are required to determine the role Ift140 plays in limb/digit formation. Mutations in IFT140 have recently been identified in Mainzer-Saldino syndrome (OMIM 266920) [55] further expanding the rapidly growing collection of human disorders caused by defects in primary cilia function.
The bfb mutant is an excellent example of a mouse model with multiple anomalies that were of interest to several investigators involved in this screen. The anomalies included blood filled blisters, isolated cleft palate, open eyelids, polydactyly and urinary tract defects. Fluid-filled or haemorrhagic blisters are the classic phenotype of the blebs mutant mice [35]. The first blebs mutant gene identified was Fras1 [56], [57] and each of the subsequently identified blebs mutations are in genes encoding structurally or functionally related proteins [35]. Mutation of FRAS1 in humans causes Fraser Syndrome which involves a series of related phenotypes to the blebs mutants [35]. The Fras1 mutation identified in the bfb mutant occurs within a structurally uncharacterised domain between the CALXβ (calcium binding) domains and the transmembrane domain. The impact of this novel mutation on FRAS1 function is unclear but given the similarity of the bfb and Fras1 null mice, it seems likely that the bfb mutation severely compromises FRAS1 function during early embryonic development.
The Lig1 mutant displayed a heritable recessive anaemic phenotype characterised by a pale liver and reduced circulating blood. The anaemic phenotype of the homozygous Lig1 mutants was associated with a relative decrease in mature RBCs and increased numbers of primitive, nucleated erythrocytes in circulating blood. This is consistent with the phenotype of the Lig1 null mutant mice described by Bentley et al., (1996) [58]. Interestingly, a human compound heterozygote carrying two mutated LIG1 alleles (Glu566Lys and Arg771Trp) has been described [59]. This patient displayed symptoms similar to Bloom syndrome, including immunodeficiency and increased cellular sensitivity to DNA-damaging agents, culminating in lethal lymphoma at age 19 [60]. Further analysis of our Lig1 mutant may reveal the molecular role that this gene plays in erythropoiesis and carcinogenesis.
We identified several pedigrees with urinary tract abnormalities with the most common anomalies being dilation of the renal pelvis and/or ureter and hydronephrosis. Three of these mutants presented with non-syndromic renal and ureter anomalies and the Fras1 mutant represented renal anomalies associated with a syndrome. These ENU mouse models are invaluable in studying the aetiology of the human condition CAKUT [61], which accounts for one-third of the anomalies detected by fetal ultrasound and can lead to end stage renal disease [62]. Whole exome sequencing is underway to identify the causal genes which will serve as candidate CAKUT genes for genetic screening of patients.
We have demonstrated that a multiple organ screen in embryonic mice performed remotely (The Australian Phenomics Facility, Canberra) to the multi-site investigators is highly feasible. In this screen, embryonic dissections were performed in a hierarchical and systematic manner (see Method S1), the technician was trained by the expert investigators to dissect and observe specific anomalies in the organs of interest. Phenotypes were recorded using digital photography and entered into the web-based, password protected database allowing all multi-site investigators to visualise phenotypes immediately. Organs and embryos were fixed and sent to the expert investigators for further analysis. The protocol we have described in detail (Method S1) forms the basis for establishing high throughput lethal screens in the future. The phenotyping protocol we outline in this report could be applicable to screening large numbers of embryonic lethal lines generated not only by ENU but also by gene targeting and gene trapping as is being undertaken by The International Knockout Mouse Consortium (IKMC). In many respects the throughput of our screen (more than 50 lines over a 2 year period) was limited not by the ability to undertake the phenotyping, but rather the generation of the ENU mutagenised pedigrees. We propose that the phenotyping protocol we describe would be easily applied to the analysis of 50–100 lines per year. Clearly this approach is not applicable to the average independent laboratory but our forecast throughput is well within the scope of existing and proposed high throughput lethal screens, including those encompassed and envisaged by the International Mouse Phenotyping Consortium (http://www.mousephenotype.org/) and the KOMP2 funding initiative (http://commonfund.nih.gov/KOMP2/). In addition, screening the same set of embryos for multiple organ anomalies provides a more comprehensive analysis of the mutant, allows the investigator to determine if they are dealing with a syndromic (as was the case with the Fras1 mutant) or non-syndromic (isolated organ anomaly) condition, is more cost effective and facilitates teamwork and collaboration.
During the time in which this screen took place there were rapid advancements in new sequencing technologies. These new technologies have subsequently superseded the need to outcross mice to an inbred strain to perform recombination mapping. For the initial mutants identified in our screen, low-resolution genome mapping was performed using SNP genotyping to identify chromosomal linkage. In the case of line 12BCC-22a, linkage to an interval on Chr 7 was identified which contained over 700 genes. Next Generation Sequencing (NGS) eliminated the need to perform further breeding, which would have been required for recombination mapping and narrowing of the interval within which to select candidates for Sanger sequencing. NGS was used to sequence all exons in this interval leading to the identification of Lig1 as the causal gene. For those mutants of interest, for which the causal genes are yet to be identified in this screen, chromosome specific or whole exome sequencing is being employed. Exome sequencing on average covers >95% of the consensus coding sequence (CCDS) exome [63]. Although mutations in non-coding regions are not isolated using exome sequencing, 99% of ENU mutations occur in splice sites and exons therefore exome sequencing should identify almost all ENU-induced mutations [7]. There will be some situations where mutations will be missed as a result of capture baits not covering specific genomic regions, the gene not being annotated as part of the CCDS, fragment mis-mapping or bad sequence coverage/quality due to high/low GC content [7], [64]. Despite these limitations gene mapping will be phased-out of future ENU-based screens and replaced with whole exome sequencing [7], [64]. Unlike recombination mapping, in which many affected and unaffected embryos are required to identify the mutated gene, exome sequencing initially only requires one affected embryo to identify the candidate casual mutation. ENU induced single nucleotide variations (SNVs) can then be subsequently validated in a larger cohort of embryos taking advantage of heritability information. This will rapidly reduce the time spent trying to identify the causative gene and allow mouse genotyping to be established earlier thereby reducing mouse husbandry costs. This will also undoubtedly decrease the percentage of valuable lines that are abandoned as a result of loss of phenotype during carrier searches and establishing heritability, as was the case in lines 12BCC-13 (lung anomalies) and 12BCC-20 (gonadal anomalies) in this study.
The efficiency of our screen at day E13.5 was 16% and at E18.5 was 33%. Many previous screens that have used ENU to identify genes that play a role during embryogenesis have focussed on earlier embryonic processes such as patterning and morphogenesis, often examining embryos at E9.5–10.5 [28], [65], [30], [66], [31], [32], or screening for post-natal lethality [11], [17], [67] and are therefore not directly comparable with our study. Additionally, some screens focused on mid-late gestation time-points but are not directly comparable because they used imaging techniques (rather than dissection) to reveal phenotypes [33], [68]. The recovery rate of mutations in our E13.5 screen was similar (but lower) to that of Ermakov et al [34] whom reported that 24% of pedigrees examined at E13.5 had consistent mutant phenotypes in two or more embryos, and our E18.5 screen was similar (but higher) to the 29% of phenotype containing pedigrees identified by dissection of E18.5 embryos by Herron et al [29]. Overall, the recovery rate reported in our study is similar to those previously reported in comparable screens.
Unfortunately, in our screen we did not recover a mutant displaying a gut phenotype. Our objective was to identify key regional gut patterning defects at E13.5 and gut epithelial defects at E18.5. The initial screening was based on identifying overall changes in morphology, length and diameter of the gastrointestinal tract in wholemounts. Although several embryos were highlighted for further examination after superficial examination, no overt anomalies were detected by histology. This indicates that attempting to screen for cellular defects based on overall morphology is not an effective method for identifying intestinal mutants. A more definitive method would be a screen based on histological criteria as was performed for the lung screen. Although feasible this is extremely labour intensive.
Conclusions
In this study we have generated ENU-induced mutant mice that model human genetic disorders. We have identified novel mutant alleles of known genes which promise to provide further insight into the function of these genes. Despite several other ENU mutagenesis screens designed to identify recessive mutations that affect embryogenesis, the genes mutated in our screen have not been previously identified in these screens. This demonstrates the high potential for the discovery of novel alleles from such screens. The associations between phenotype and genotype which can be identified without prior gene bias, using ENU mutagenesis are invaluable in providing insight into the mechanisms that underlie normal development and human congenital disease.
Methods
ENU-mutagenesis
Animal experiments were conducted with approval of the Australian National University Animal Experimentation Ethics Committee and carried out in accordance with institutional guidelines. Male C57BL/6J mice were given three weekly intraperitoneal injections of 90 mg ENU/kg body weight as described previously [69]. After an 8 week recovery period the ENU treated male mice (G0) were crossed with either C57BL/6 or C3H/HeH (C3H) females to generate 89 founder males (F1). To screen for recessive mutations a standard three generation breeding protocol was used (Figure 1). F1 males which were heterozygous for the ENU induced mutations were crossed to C57BL/6J or C3H females to generate G2 progeny. The G2 daughters, half of which were heterozygous for the mutations, were backcrossed to their F1 fathers to generate G3 embryos that were screened at E13.5 and/or E18.5 for anomalies in skin, lung, kidney, ureter, vasculature, blood, gut, gonad and limb/craniofacial structures. Biopsies were taken from the embryos or yolk sacs for DNA extraction. A minimum of 4 litters were screened (average litter size 6–8 embryos and a minimum of 24 embryos) per pedigree and time-point. 12.5% of the G3 embryos were expected to be homozygous for the mutations. 52 lines were screened to completion with 37 additional lines abandoned due to breeding issues and small litter sizes.
Screen
G3 embryos were examined for gross external visible anomalies of the skin, limb and craniofacial structures, haemorrhages, oedema (vascular abnormalities) and anaemia (blood abnormalities). Embryos were then dissected and lungs, gut, kidneys, ureters and gonads were microscopically examined. Blood, vascular and gonadal anomalies were examined microscopically at E13.5 only. Gonads were screened for ovotestes and abnormalities in testis cord formation (eg smaller gonads, fewer or disorganised cords). Lungs were examined histologically at E18.5 only. Skin, limb/craniofacial structures, kidneys, ureters and gut were examined at both time points. Anomalies were recorded using digital photography. Results and photographs were entered in a web-based, password-protected, in-house database, Musterer. All carcasses and organs were fixed in 10% formalin. A full description of the screening protocol is provided in the Supplementary Information (Method S1).
Sex Genotyping of Mouse Embryos
Embryos were sexed by PCR using primers based on the mouse Smcx and Smcy genes as previously described [70]. Genomic DNA was isolated from the yolk sacs of embryos exhibiting testis anomalies for sex genotyping.
Histology
All fixed E18.5 lungs were embedded in paraffin and 5 µm sections were stained with haematoxylin and eosin. Sections were examined for defects in distal lung development including condensed (thickened) lung mesenchyme, dilated distal airspaces, overt defects in vascular development and abnormal progression from pseudostratified columnar epithelial cells lining proximal airspaces to cuboidal/squamous epithelial cells lining the distal airspaces.
Blood Analysis
Cytospins were performed on whole blood at 500×g for 4 minutes in a Cytospin 3 (Shandon). Cells were fixed in 100% methanol and stained with May-Grunwald-Giemsa.
DNA Isolation
DNA was extracted from embryonic tissue using proteinase K digestion and precipitation. Approximately 50 mg of tissue was lysed at 55°C for 4 hours in 500 µl buffer containing 50 mM Tris.HCl, pH 8 (Sigma), 100 mM EDTA (Sigma), 0.5% SDS (Sigma) and 200 µg/ml proteinase K (Bioline). Following centrifugation at 18000×g for 10 min the supernatant was mixed with 150 µl 5 M NaCl (Sigma) and incubated at room temperature for 10 min. The supernatant from a second centrifugation (as previously) was mixed with 450 µl ice-cold isopropanol. The DNA was pelleted by a third centrifugation step and washed twice in 100 µl 70% ethanol prior to drying and resuspending in 100 µl of Tris.HCl buffered water. The quality and concentration of the DNA was determined by spectrophotometer absorbance readings at 260, 280 and 230 nm, followed by agarose gel electrophoresis analysis.
Mapping
For linkage analysis genomic DNA from approximately 10–20 affected and 10–20 unaffected G3 C57BL/6J×C3H mice were screened with 41 strain specific SNP markers at proximal, distal and central positions on each chromosome using the Amplifluor SNP genotyping system (Chemicon, Millipore). Additional SNP markers were used within linked regions to further fine map the causal mutation in each strain.
Sequencing
Sequencing of candidate genes was performed to locate the causal base substitution. DNA was prepared from an individual affected mouse. Primers were designed for candidate genes to amplify all exons +/−15 bp to cover splice junctions. Amplicons were then Sanger sequenced on an Applied Biosystems 3730xl capillary sequencer. This automated platform uses Big Dye Terminator (BDT) chemistry version 3.1 (Applied Biosystems). The raw trace files were analysed using Lasergene software (DNAstar) against the C57BL/6J mouse reference genome (mm9/NCBI m37).
Next Generation Sequencing
A SureSelect custom solution array (Agilent) was designed using the online tool, eArray to include Refseq release 43 (NCBI) annotated exons (plus splice donor and acceptor sites) within the linkage interval. 100 bp paired end Illumina libraries of the captured regions from a single affected mouse were produced and run in a single lane of an Illumina GAIIx. Sequence reads were mapped to the NCBIM37 assembly of the reference mouse genome using the bowtie aligner47. Untrimmed reads were aligned allowing a maximum of two sequence mismatches and were discarded where they aligned to the genome more than once. Sequence variants were identified with SAMtools and custom perl scripts were subsequently used to identify those which occurred within exons and splice donor/acceptor sites, and were not known or strain-specific variants.
Mutation Validation
Single nucleotide variations (SNVs) identified by Sanger or Next Generation Sequencing were validated using the Amplifluor SNP genotyping system (Chemicon, Millipore). Assays were designed to each SNV of interest and validated against a set of embryos affected for the phenotype of interest and a set of unaffected embryos. Amplifluor validation primers for Fras1, Ift140 and Lig1 are listed in Table S1. For the bfb line 39 embryos (26 unaffected and 13 affected), for the cauli line 37 embryos (29 unaffected and 8 affected) and for the Lig1 line 20 embryos (10 affected and 10 unaffected) were genotyped to confirm the association of genotype to phenotype. In all cases the genotype correlated with the phenotype.
Immunofluorescence
Testes were fixed, processed and sectioned as described previously [71]. For indirect immunofluorescence staining, frozen sections were incubated with Anti-AMH goat polyclonal antibody (1∶200; Santa Cruz, sc-6886) followed by detection with an Alexa dye-linked secondary antibody (1∶1000; Molecular Probes). Images were captured using fluorescence microscopy (Olympus Corp., NY, USA).
Supporting Information
Amplifluor Genotyping Primers.
(DOC)
ENU Organogenesis Screening Protocol.
(DOC)
Acknowledgments
We would like to thank the staff of the Australian Phenomics Facility at The Australian National University, particularly Heather Wilson, Vicki Adams, Robert Tunningley, Kartik Saxena and Allister Leong, who skilfully dissected the embryos and made the initial phenotypic observations, and Jessica Armstrong and Alanna Craven for their project management. We would also like to thank Edward Bertram for introducing us to the ENU resources available at the Australian Phenomics Facility. We would also like to thank Alison Moxham from The Ritchie Centre, Monash Institute of Medical Research, Monash University for assistance with lung histology.
Funding Statement
This work was enabled by the Australian Phenomics Network and partly supported by funding from the Australian Government’s National Collaborative Research Infrastructure Strategy, a Strategic Grant from the Faculty of Medicine, Nursing and Health Sciences at Monash University, and the Victorian Government’s Operational Infrastructure Support Program. IS acknowledges support through the NH&MRC R. Douglas Wright and ARC Future Fellowship schemes. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. O’Sullivan G, O’Tuathaigh CM, Clifford JJ, O’Meara GF, Croke DT, et al. (2006) Potential and limitations of genetic manipulation in animals. Drug Discovery Today: Technologies. 3: 173–180. [DOI] [PubMed] [Google Scholar]
- 2. Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A (1999) Mouse ENU mutagenesis. Hum Mol Genet. 8: 1955–63. [DOI] [PubMed] [Google Scholar]
- 3. Anderson KV (2000) Finding the genes that direct mammalian development: ENU mutagenesis in the mouse. Trends Genet. 16: 99–102. [DOI] [PubMed] [Google Scholar]
- 4. Clark AT, Goldowitz D, Takahashi JS, Vitaterna MH, Siepka SM, et al. (2004) Implementing large-scale ENU mutagenesis screens in North America. Genetica. 122: 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takahasi KR, Sakuraba Y, Gondo Y (2007) Mutational pattern and frequency of induced nucleotide changes in mouse ENU mutagenesis. BMC Mol Biol. 8: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nguyen N, Judd LM, Kalantzis A, Whittle B, Giraud AS, et al. (2011) Random mutagenesis of the mouse genome: a strategy for discovering gene function and the molecular basis of disease. Am J Physiol Gastrointest Liver Physiol. 300: G1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Andrews TD, Whittle B, Field MA, Balakishnan B, Zhang Y, et al. (2012) Massively parallel sequencing of the mouse exome to accurately identify rare, induced mutations: an immediate source for thousands of new mouse models. Open Biol 2: 120061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Z, Alpert D, Francis R, Chatterjee B, Yu Q, et al. (2009) Massively parallel sequencing identifies the gene Megf8 with ENU-induced mutation causing heterotaxy. Proc Natl Acad Sci U S A. 106: 3219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sabrautzki S, Rubio-Aliaga I, Hans W, Fuchs H, Rathkolb B, et al. (2012) New mouse models for metabolic bone diseases generated by genome-wide ENU mutagenesis. Mamm Genome. 23: 416–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun M, Mondal K, Patel V, Horner VL, Long AB, et al. (2012) Multiplex Chromosomal Exome Sequencing Accelerates Identification of ENU-Induced Mutations in the Mouse. G3 (Bethesda) 2: 143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kile BT, Hentges KE, Clark AT, Nakamura H, Salinger AP, et al. (2003) Functional genetic analysis of mouse chromosome 11. Nature. 425: 81–6. [DOI] [PubMed] [Google Scholar]
- 12. Clark AT, Firozi K, Justice MJ (2004) Mutations in a novel locus on mouse chromosome 11 resulting in male infertility associated with defects in microtubule assembly and sperm tail function. Biol Reprod. 70: 1317–24. [DOI] [PubMed] [Google Scholar]
- 13. Hentges KE, Nakamura H, Furuta Y, Yu Y, Thompson DM, et al. (2006) Novel lethal mouse mutants produced in balancer chromosome screens. Gene Expr Patterns. 6: 653–65. [DOI] [PubMed] [Google Scholar]
- 14. Boles MK, Wilkinson BM, Maxwell A, Lai L, Mills AA, et al. (2009) A mouse chromosome 4 balancer ENU-mutagenesis screen isolates eleven lethal lines. BMC Genet. 10: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ching YH, Munroe RJ, Moran JL, Barker AK, Mauceli E, et al. (2010) High resolution mapping and positional cloning of ENU-induced mutations in the Rw region of mouse chromosome 5. BMC Genet. 11: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hagarman JA, O’Brien TP (2009) An essential gene mutagenesis screen across the highly conserved piebald deletion region of mouse chromosome 14. Genesis. 47: 392–403. [DOI] [PubMed] [Google Scholar]
- 17. Bogani D, Willoughby C, Davies J, Kaur K, Mirza G, et al. (2005) Dissecting the genetic complexity of human 6p deletion syndromes using a region-specific, phenotype-driven mouse screen. Proc Natl Acad Sci U S A 102: 12477–12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buac K, Watkins-Chow DE, Loftus SK, Larson DM, Incao A, et al. (2008) A Sox10 expression screen identifies an amino acid essential for Erbb3 function. PLoS Genet. 2008 4): e1000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matera I, Watkins-Chow DE, Loftus SK, Hou L, Incao A, et al. (2008) A sensitized mutagenesis screen identifies Gli3 as a modifier of Sox10 neurocristopathy. Hum Mol Genet. 17: 2118–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stottmann RW, Moran JL, Turbe-Doan A, Driver E, Kelley M, et al. (2011) Focusing forward genetics: a tripartite ENU screen for neurodevelopmental mutations in the mouse. Genetics. 188: 615–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anderson NM, Berberovic Z, Berndl E, Bailey ML, Flenniken AM, et al. (2012) Cytopenia induction by 5-fluorouracil identifies thrombopoietic mutants in sensitized ENU mutagenesis screens. Exp Hematol. 40: 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rastan S, Hough T, Kierman A, Hardisty R, Erven A, et al. (2004) Towards a mutant map of the mouse-new models of neurological, behavioural, deafness, bone, renal and blood disorders Genetica. 122: 47–9.23. [DOI] [PubMed] [Google Scholar]
- 23. Aigner B, Rathkolb B, Herbach N, Kemter E, Schessl C, et al. (2007) Screening for increased plasma urea levels in a large-scale ENU mouse mutagenesis project reveals kidney disease models. Am J Physiol Renal Physiol. 292: F1560–7. [DOI] [PubMed] [Google Scholar]
- 24. Hoyne GF, Goodnow CC (2006) The use of genomewide ENU mutagenesis screens to unravel complex mammalian traits: identifying genes that regulate organ-specific and systemic autoimmunity. Immunol Rev. 210: 27–39. [DOI] [PubMed] [Google Scholar]
- 25. Kennedy CL, O’Connor AE, Sanchez-Partida LG, Holland MK, Goodnow CC, et al. (2006) A repository of ENU mutant mouse lines and their potential for male fertility research. Mol Hum Reprod. 11: 871–80. [DOI] [PubMed] [Google Scholar]
- 26. Srivastava AK, Mohan S, Wergedal JE, Baylink DJ (2003) A genomewide screening of N-ethyl-N-nitrosourea-mutagenized mice for musculoskeletal phenotypes. Bone. 33: 179–91. [DOI] [PubMed] [Google Scholar]
- 27.Brown FC, Scott N, Rank G, Collinge JE, Vadolas J, et al. (2012) ENU mutagenesis identifies the first mouse mutants reproducing human β-thalassemia at the genomic level. Blood Cells Mol Dis. 2012 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 28. Kasarskis A, Manova K, Anderson KV (1998) A phenotype-based screen for embryonic lethal mutations in the mouse. Proc Natl Acad Sci U S A. 95: 7485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Herron BJ, Lu W, Rao C, Liu S, Peters H, et al. (2002) Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat Genet 30: 185–9. [DOI] [PubMed] [Google Scholar]
- 30. García-García MJ, Eggenschwiler JT, Caspary T, Alcorn HL, Wyler MR, et al. (2005) Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc Natl Acad Sci U S A 102: 5913–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wansleeben C, van Gurp L, Feitsma H, Kroon C, Rieter E, et al. (2011) An ENU-mutagenesis screen in the mouse: identification of novel developmental gene functions. PLoS One. 20116: e19357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sandell LL, Iulianella A, Melton KR, Lynn M, Walker M, et al. (2011) A Phenotype-Driven ENU Mutagenesis Screen Identifies Novel Alleles With Functional Roles in Early Mouse Craniofacial Development. Genesis. 49: 342–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu Q, Shen Y, Chatterjee B, Siegfried BH, Leatherbury L, et al. (2004) ENU induced mutations causing congenital cardiovascular anomalies. Development. 131: 6211–23. [DOI] [PubMed] [Google Scholar]
- 34. Ermakov A, Stevens JL, Whitehill E, Robson JE, Pieles G, et al. (2009) Mouse mutagenesis identifies novel roles for left-right patterning genes in pulmonary, craniofacial, ocular, and limb development. Dev Dyn. 238: 581–94. [DOI] [PubMed] [Google Scholar]
- 35. Smyth I, Scambler P (2005) The genetics of Fraser syndrome and the blebs mouse mutants. Hum Mol Genet 2: R269–74. [DOI] [PubMed] [Google Scholar]
- 36. Ashe A, Butterfield NC, Town L, Courtney AD, Cooper AN, et al. (2012) Mutations in mouse Ift144 model the craniofacial, limb and rib defects in skeletal ciliopathies. Human Molecular Genetics 21: 1808–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liem KF Jr, Ashe A, He M, Satir P, Moran J, et al. (2012) The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. The Journal of Cell Biology 197: 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pedersen LB, Rosenbaum JL (2008) Chapter Two Intraflagellar Transport (IFT): Role in Ciliary Assembly, Resorption and Signalling. Current Topics in Developmental Biology, 85, 23–61. [DOI] [PubMed] [Google Scholar]
- 39. Behal RH, Miller MS, Qin H, Lucker BF, Jones A, et al. (2012) Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins.J Biol Chem. 287: 11689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takahashi S, Onodera K, Motohashi H, Suwabe N, Hayashi N, et al. (1997) Arrest in primitive erythroid cell development caused by promoter-specific disruption of the GATA-1 gene. J Biol Chem 272: 12611–5. [DOI] [PubMed] [Google Scholar]
- 41. Tsang AP, Fujiwara Y, Hom DB, Orkin SH (1998) Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev 12: 1176–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T (2004) Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature, 432; 473–478. [DOI] [PubMed] [Google Scholar]
- 43. Wilhelm D, Palmer S, Koopman P (2007) Sex determination and gonadal development in mammals. Physiol Rev. 87: 1–28. [DOI] [PubMed] [Google Scholar]
- 44.Munger SC, Aylor DL, Syed HA, Magwene PM, Threadgill DW, et al. (2009) Elucidation of the transcription network governing mammalian sex determination by exploiting strain-specific susceptibility to sex reversal Genes Dev 23, 2521–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ichikawa I, Kuwayama F, Pope JC 4th, Stephens FD, Miyazaki Y (2002) Paradigm shift from classic anatomic theories to contemporary cell biological views of CAKUT. Kidney Int 61: 889–98. [DOI] [PubMed] [Google Scholar]
- 46. Gimpel C, Masioniene L, Djakovic N, Schenk JP, Haberkorn U, et al. (2010) Complications and long-term outcome of primary obstructive megaureter in childhood. Pediatr Nephrol 25: 1679–86. [DOI] [PubMed] [Google Scholar]
- 47. Kerecuk L, Schreuder MF, Woolf AS (2008) Renal tract malformations: perspectives for nephrologists. Nat Clin Pract Nephrol 4: 312–25. [DOI] [PubMed] [Google Scholar]
- 48.Hooper SB, Wallace MJ (2004) Chapter 9: Role of physical, endocrine and growth factors in lung development. In: The Lung: Development, Aging and the Environment. Ed. R. Harding, KE. Pinkerton & CG. Plopper. Elsevier Academic Press, London UK. p131–148. [Google Scholar]
- 49. Hooper SB, Wallace MJ (2006) Role of physico-chemical environment in lung development. Clinical and Experimental Pharmacology and Physiology 33: 273–279. [DOI] [PubMed] [Google Scholar]
- 50. Cole TJ, Blendy JA, Monaghan AP, Krieglstein K, Schmid W, et al. (1995) Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev 9: 1608–1621. [DOI] [PubMed] [Google Scholar]
- 51.WHO (2003) Global registry and database on craniofacial anomalies: Chapter2; Craniofacial anomalies and associated birth defects; WHO registry meeting on craniofacial anomalies, Bauru, Brazil. [Google Scholar]
- 52. Cameron DA, Pennimpede T, Petkovich M (2009) Tulp3 is a critical repressor of mouse hedgehog signalling. Dev Dynamics 238: 1140–9. [DOI] [PubMed] [Google Scholar]
- 53. Patterson VL, Damrau C, Paudyal A, Reeve B, Grimes DT, et al. (2009) Mouse hitchhiker mutants have spina bifida, dorso-ventral patterning defects and polydactyly: identification of Tulp3 as a novel negative regulator of the Sonic hedgehog pathway. Hum Mol Genet. 18: 1719–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Milenkovic L, Goodrich LV, Higgins KM, Scott MP (1999) Mouse patched1 controls body size determination and limb patterning. Development. 126: 4431–40. [DOI] [PubMed] [Google Scholar]
- 55. Perrault I, Saunier S, Hanein S, Filhol E, Bizet AA, et al. (2012) Mainzer-Saldino syndrome is a ciliopathy caused by IFT140 mutants. Am J Hum Genet. 90: 864–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vrontou S, Petrou P, Meyer BI, Galanopoulos VK, Imai K, et al. (2003) Fras1 deficiency results in cryptophthalmos, renal agenesis and blebbed phenotype in mice. Nat Genet 34: 209–14. [DOI] [PubMed] [Google Scholar]
- 57. McGregor L, Makela V, Darling SM, Vrontou S, Chalepakis G, et al. (2003) Fraser syndrome and mouse blebbed phenotype caused by mutations in FRAS1/Fras1 encoding a putative extracellular matrix protein. Nat Genet 34: 203–8.33. [DOI] [PubMed] [Google Scholar]
- 58. Bentley DJ, Selfridge J, Millar JK, Samuel K, Hole N, et al. (1996) DNA ligase I is required for foetal liver erythropoiesis but is not essential for mammalian cell viability. Nat. Genet. 13: 489–491. [DOI] [PubMed] [Google Scholar]
- 59.Barnes DE, Tomkinson AE, Lehmann AR, Webster ADB, Lindahl T (1992) Mutations in the DNA ligase I gene in an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell 69, 495–504. [DOI] [PubMed] [Google Scholar]
- 60. Webster AD, Barnes DE, Arlett CF, Lehmann AR, Lindahl T (1992) Growth retardation and immunodeficiency in a patient with mutations in the DNA ligase I gene. Lancet 339: 1508–9. [DOI] [PubMed] [Google Scholar]
- 61. Miyazaki Y, Ishikawa I (2003) Ontogeny of congenital anomalies of the kidney and urinary tract, CAKUT. Pediatr Int 45: 598–604. [DOI] [PubMed] [Google Scholar]
- 62. Woolf AS, Price KL, Scambler PJ, Winyard PJ (2004) Evolving concepts in human renal dysplasia J Am Soc Nephrol. 15: 998–1007. [DOI] [PubMed] [Google Scholar]
- 63. Fairfield H, Gilbert GJ, Barter M, Corrigan RR, Curtain M, et al. (2011) Mutation discovery in mice by whole exome sequencing Genome Biol. 12: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Benjamini Y, Speed TP (2012) Summarizing and correcting the GC content bias in high-throughput sequencing Nucleic Acids Research. 40: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bogani D, Warr N, Elms P, Davies J, Tymowska-Lalanne Z, et al. (2004) New semidominant mutations that affect mouse development. Genesis. 40: 109–117. [DOI] [PubMed] [Google Scholar]
- 66. Zohn IE, Anderson KV, Niswander L (2005) Using genomewide mutagenesis screens to identify the genes required for neural tube closure in the mouse. Birth Defects Res A Clin Mol Teratol. 73: 583–90. [DOI] [PubMed] [Google Scholar]
- 67. Kamp A, Peterson MA, Svenson KL, Bjork BC, Hentges KE, et al. (2010) Genome-wide identification of mouse congenital heart disease loci. Hum Mol Genet. 19: 3105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pieles G, Geyer SH, Szumska D, Schneider J, Neubauer S, et al. (2007) microMRI-HREM pipeline for high-throughput, high-resolution phenotyping of murine embryos. J Anat. 211: 132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hitotsumachi S, Carpenter DA, Russell WL (1985) Dose-repetition increases the mutagenic effectiveness of N-ethyl-N-nitrosourea in mouse spermatogonia. Proc Natl Acad Sci USA 82: 6619–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Barrionuevo F, Bagheri-Fam S, Klattig J, Kist R, Taketo MM, et al. (2006) Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol Reprod. 74(1): 195–201. [DOI] [PubMed] [Google Scholar]
- 71. Bagheri-Fam S, Argentaro A, Svingen T, Combes AN, Sinclair AH, et al. (2011) Defective survivial of proliferating Sertoli cells and androgen receptor function in a mouse model of the ATR-X syndrome. Hum Mol Genet. 20: 2213–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amplifluor Genotyping Primers.
(DOC)
ENU Organogenesis Screening Protocol.
(DOC)