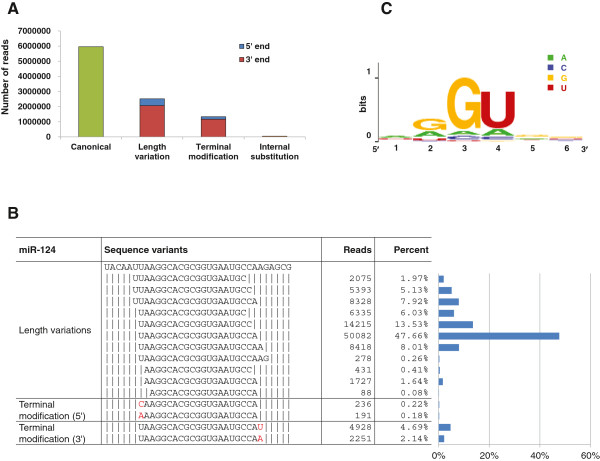
Figure 4.
miRNA sequence variants. (A) Distribution of sequence variation types among all miRNA reads. (B) Sequence variants in miR-124. The nucleotides differing from the template genome are highlighted in red. Blue bars to the right indicate frequencies of these variants. (C) Motifs identified among internal substitution sites. The overall height of a stack indicates sequence conservation at that position: the higher the stack, the more the position is conserved. The heights of nucleotides within a stack indicate the relative frequency for each nucleotide at that position. We analyzed 6-nucleotide sequences containing an internal substitution site having a substitution rate > 5%, and found that a GGU motif was preferentially present among all sequences. The numbers (1–6) below the X axis indicate nucleotide positions in the motifs.