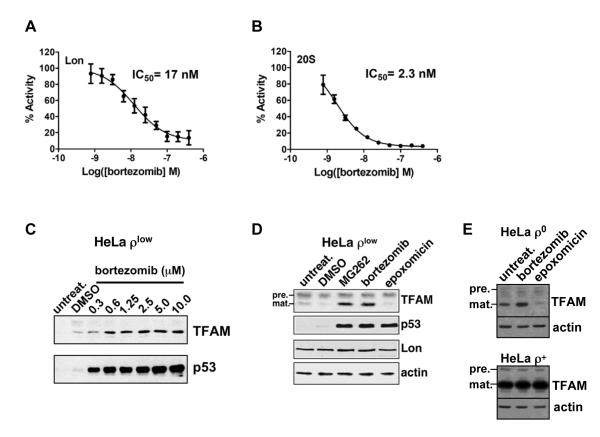
Figure 3. Lon-dependent proteolysis of TFAM is blocked by bortezomib and MG262 but not epoxomicin.
(A) and (B) Lon (200 nM monomer) or 20S (3 nM complex) peptidase activities were measured using the fluorescent dipeptide substrate AA2-Rh110 (6 μM) incubated in the presence or absence of bortezomib at 37°C for 3 hr. Fluorescence was normalized to percent activity of no drug control. Results represent at least 3 independent experiments. (C) ρlow cells were incubated with or without bortezomib for 18 hr and extracts were blotted for TFAM or p53. (D) and (E) ρlow, ρ0 or ρ+ cells were treated with DMSO, bortezomib (5 μM), MG262 (1.25 μM) or epoxomicin (1 μM) for 18 hr; extracts were blotted for TFAM, p53, Lon or actin. TFAM precursor (pre.) and mature (mat.) proteins are indicated.