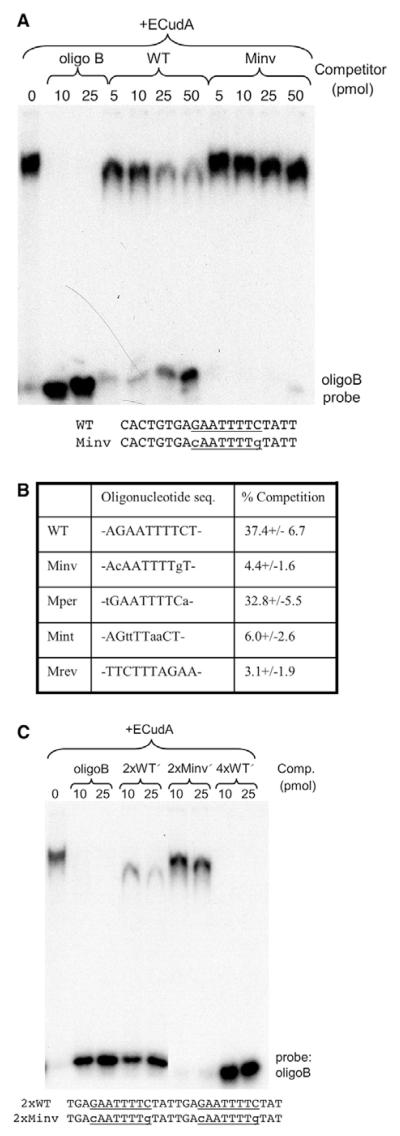
Fig. 5. Mutational analysis of ECudA binding to the cotC dyad.
(A) The WT 20-mer competitor oligonucleotide (CACTGTGAGAATTTTCTATT) encompasses the interrupted dyad (underlined); the C and G within the dyad sequence are swapped in the Minv competitor. WT and Minv were used in band shift assays with oligonucleotide B and ECudA. The assays were performed using Cy5-labelled probes and the intensity of the unshifted probe bands and of the shifted (retarded) bands was measured. (B) The competition efficiency with 100 pmol of competitor is calculated as the ratio, in percentage terms, of the retarded signal to the total (retarded + unretarded) signal. This is a compilation of data from three experiments and the percentage is expressed ±s.d. (C) Point mutation analysis of ECudA binding to multimeric forms of the dyad using the dimeric competitor oligonucleotides, WT′ and Minv′, that contain tandem repeats of the 14-mer encompassing the dyad sequence. A tetramer of the WT′ form (4×WT′) was also used that contains two tandemly arrayed copies of the dimer.