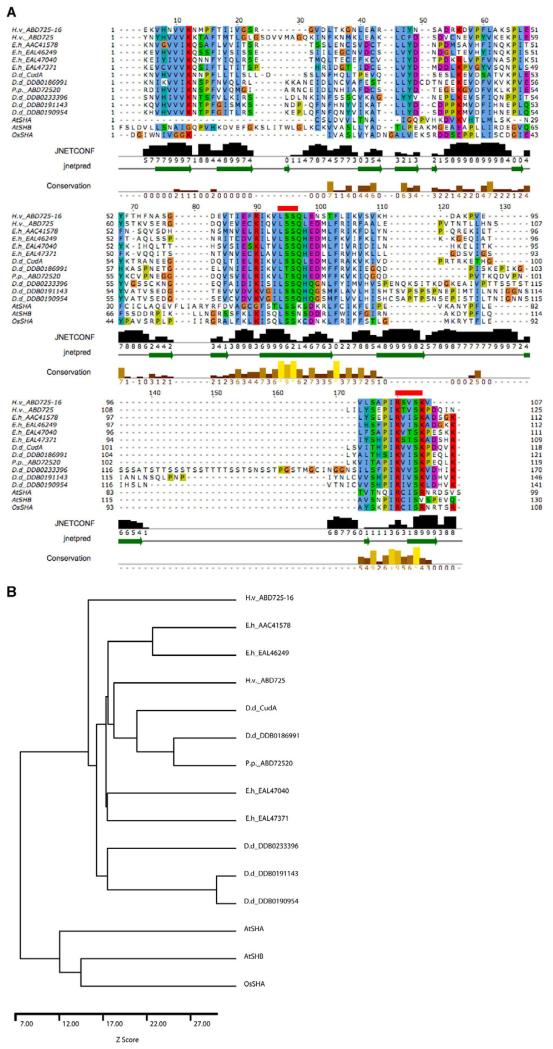
Fig. 9. CudA homologues and the evolution of STAT proteins.
(A) The core region of the Dictyostelium CudA protein compared with related proteins. Amoebozoan species of origin: D.d., Dictyostelium discoideum; E.h., Entamoeba histolytica; H.v., Hartmanella vermiformis; P.p., Physarum polycephalum. The alignment of the core domain of CudA with proteins of similar sequence was produced by AMPS (Barton, 1994), followed by manual adjustment in Jalview (Clamp et al., 2004). Accession numbers are shown only for those proteins for which no publication is available; ECudA is AAC41578. The alignment is coloured according to the ClustalX colour scheme (Thompson, et al., 1997). Residues are coloured by their physico-chemical properties as well as by how frequently they occur at each position. Thus, residues are only coloured if they show similarity to a notional ‘consensus’. Negatively charged residues are in purple; hydrophobic residues in blue; positively charged residues in red; and polar residues in green. Since glycine and proline have special properties, they are separately coloured in orange and mustard, respectively. The secondary structure prediction produced by JPred/JNet (Cuff and Barton, 2000) is shown below the alignment. The green arrows within the ‘jnetpred’ line represent predicted β-strands. The bar chart and numbers labelled ‘JNETCONF’ show the prediction confidence on a scale of 0-9. The ‘Conservation’ line highlights positions in the alignment where the physico-chemical properties of the amino acids are most highly conserved. The two red lines above the sequence show the positions of the mutations introduced to assess the importance of the two regions in DNA binding. (B) Dendrogram for the sequences shown in A. The sequences were compared pairwise and a Z-score calculated from 100 randomisations using AMPS (Barton and Sternberg, 1987).