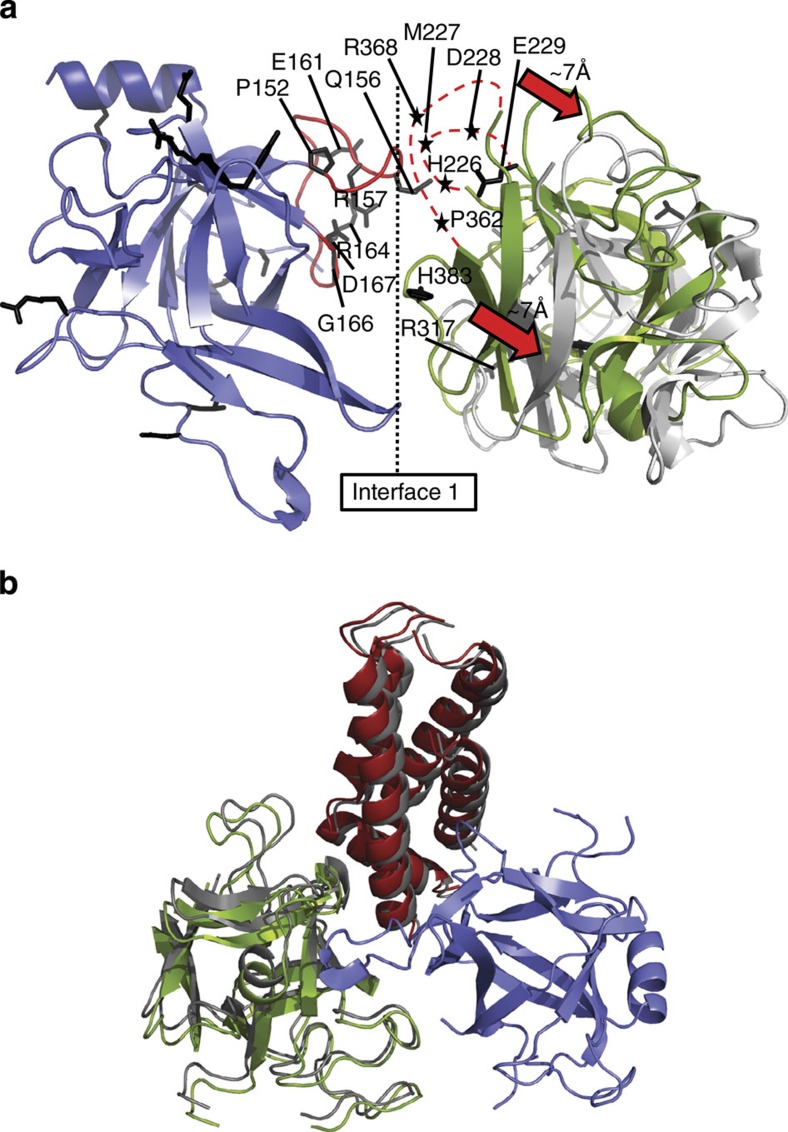
Figure 2. Relative positions between and within N-terminal disease hot spots.
(a) Superposition of domain A (blue) in the open- and closed-state models, showing the relative position of domain B in a neighbouring subunit (closed: green and open: grey). Disease mutations are shown as black sticks. Select mutations in three juxtaposed loops are labelled. The β8–β9 loop in domain A is shown in red. Two flexible loops in domain B that contain multiple disease mutations are shown as red dotted lines, with disease mutations indicated by black asterisks. This interface is subject to a total of 20 disease mutations: P152S*, Q156K, R157Q*, E161G, R164C, R164L, R164Q*, G166R, D167N, D167G, H226R*, M227K, D228V, E229K*, R317L, R317W*, P362M*, R368L, R368Q and H383N, where asterisks indicate mutations found in RyR2 (all in rabbit RyR1 residue numbering). (b) Superposition of the closed (colours) and open (grey) RyR1ABC models, based on domain A (blue), indicating the relative positions of domains B (green) and C (red). These conformational changes are too small to be interpreted at the resolution of the cryoEM maps, so channel opening does not seem to induce major conformational rearrangements within each hot spot.