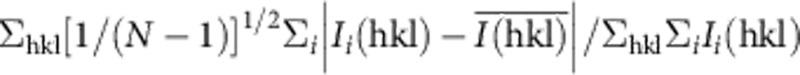Table 1. Crystallographic parameters.
| L14R | C36R | R45C | D61N | V219I | G249R | R402G | I404M | RyR1BC | |
|---|---|---|---|---|---|---|---|---|---|
| Data collection | |||||||||
| Space group | R32 | R32 | P6322 | R32 | R32 | R32 | P6322 | R32 | P3121 |
| Cell dimensions | |||||||||
| a=b, c (Å) | 171.40, 300.48 | 170.38, 302.11 | 118.16, 246.95 | 168.63, 301.65 | 169.27, 305.60 | 167.49, 304.70 | 118.26, 243.93 | 171.37, 299.43 | 68.45, 131.89 |
| Resolution (Å) | 50-2.9 (3.06-2.90) | 50-2.8 (2.95-2.80) | 50-2.5 (2.64-2.50) | 50-2.95 (3.11-2.95) | 50-2.8 (2.95-2.80) | 50-2.4 (2.53-2.40) | 50-2.95 (3.11-2.95) | 50-2.5 (2.64-2.50) | 50-2.73 (2.80-2.73) |
| Rsym or Rmerge* | 14.7 (212.7) | 11.1 (152.9) | 16.0 (151.2) | 15.0 (151.5) | 12.9 (130.9) | 6.7 (100.0) | 12.9 (64.5) | 9.7 (101.9) | 13.0 (81.4) |
| Rpim† | 3.1 (45.3) | 3.4 (47.1) | 5.0 (46.4) | 4.6 (46.6) | 4.5 (45.9) | 3.1 (46.5) | 7.4 (37.2) | 3.7 (39.3) | 4.1 (25.5) |
| I/σI | 14.6 (1.8) | 18.0 (1.9) | 13.4 (1.9) | 12.6 (1.8) | 15.6 (2.0) | 16.0 (1.8) | 10.0 (2.1) | 12.9 (1.9) | 15.5 (3.4) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.00) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) | 99.8 (100.0) | 100.0 (100.0) | 100 (100.0) |
| Redundancy | 22.7 (22.8) | 11.4 (11.5) | 10.9 (11.1) | 11.4 (11.5) | 9.1 (9.1) | 5.7 (5.7) | 3.9 (4.0) | 7.6 (7.6) | 10.8 (11.1) |
| Refinement | |||||||||
| Resolution (Å) | 50-2.9 | 50-2.8 | 50-2.5 | 50-2.95 | 50-2.8 | 50-2.4 | 50-2.95 | 50-2.5 | 50-2.73 |
| Number of reflections | 35972 | 39670 | 34297 | 33250 | 39637 | 61106 | 20871 | 55655 | 9595 |
| Rwork/Rfree | 22.8/24.9 | 24.3/24.6 | 26.0/27.7 | 22.7/25.7 | 23.5/25.3 | 23.9/25.8 | 25.1/28.9 | 23.2/24.9 | 22.4/24.1 |
| Number of atoms | |||||||||
| Protein | 3521 | 3495 | 3297 | 3546 | 3575 | 3603 | 3409 | 3598 | 1968 |
| Ligand/ion | 6 | 6 | 0 | 6 | 6 | 0 | 0 | 6 | 0 |
| Water | 16 | 17 | 56 | 14 | 36 | 99 | 27 | 70 | 16 |
| B-factors (Å2) | |||||||||
| Protein | 61.7 | 24.1 | 22.3 | 39.2 | 31.3 | 31.5 | 18.9 | 21.4 | 26.4 |
| Ligand/ion | 132.7 | 104.2 | n/a | 91.1 | 107.8 | n/a | n/a | 95.7 | n/a |
| Water | 49.5 | 21.6 | 20.3 | 26.8 | 21.9 | 28.3 | 13.2 | 21.0 | 18.9 |
| RMSD | |||||||||
| Bond lengths (Å) | 0.006 | 0.006 | 0.005 | 0.006 | 0.005 | 0.005 | 0.007 | 0.006 | 0.011 |
| Bond angles (°) | 1.013 | 0.975 | 0.902 | 0.992 | 0.902 | 0.892 | 1.004 | 0.942 | 1.314 |
RMSD, root mean square deviation.Values in parentheses are for highest-resolution shell.
*Merging R factor: Rmerge=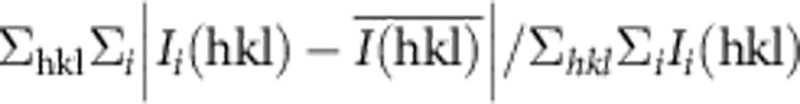
†Precision-indicating merging R factor: Rpim=