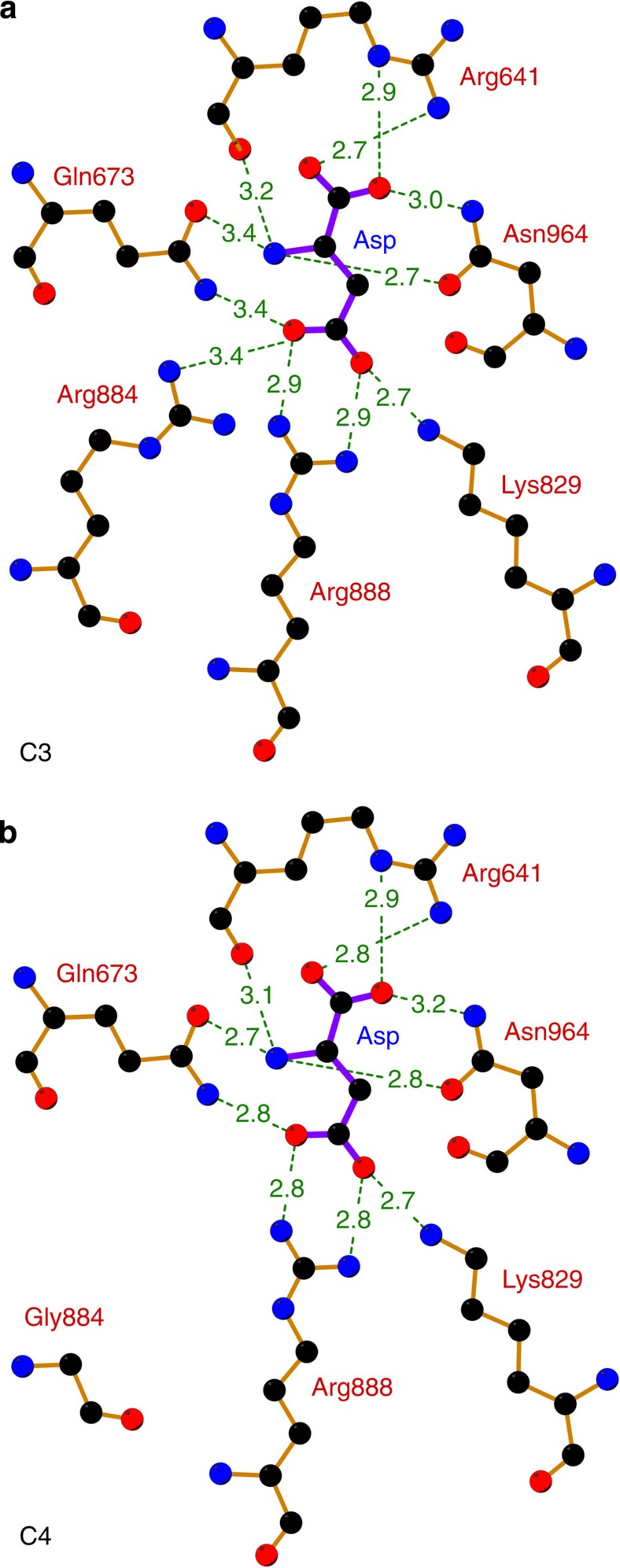
Figure 4. Side by side comparison of the feedback inhibitor-binding site.
(a) Inhibitor-binding site of C3 PEPC (F. pringlei). (b) Inhibitor-binding site of C4 PEPC (F. trinervia). The distances between the inhibitor aspartate and amino acids of the binding pocket are given in Å. The estimated coordinate errors are 0.24 Å (F. pringlei C3 PEPC) and 0.29 Å (F. trinervia C4 PEPC). Residues Arg641, Lys829, Arg888 and Asn964 have been identified as the malate-binding motif15. In C3 PEPC, Arg884 provides an additional hydrogen bond for inhibitor binding. In C4 PEPC, this residue is replaced by glycine, which is 6.9 Å away from the inhibitor molecule.