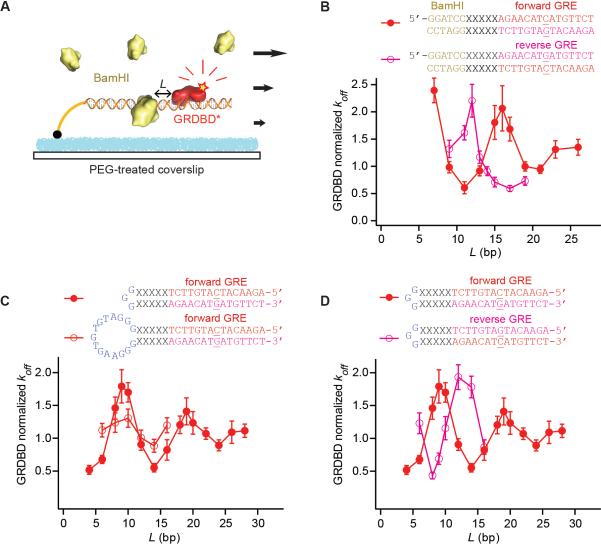
Fig. 1.
Allostery through DNA affecting koff of GRDBD near BamHI or near a hairpin loop. (A) Schematic for the single-molecule assay in a flow cell. The structural model is for L = 11 with GRDBD from Protein Data Bank (PDB) ID 1R4R (13) and BamHI from PDB ID 2BAM (32). (B) Oscillation in the koff of GRDBD for the forward (red solid circles) and reverse (magenta open circles) GRE sequences, normalized to that measured in the absence of BamHI (±SEM). DNA sequences are shown with the linker DNA (L = 5). The central base of GRE, which makes the sequence non-palindromic, is underlined. (C) Protein binding affinity affected by a nearby DNA hairpin loop, 3 bp and 15 bp (±SEM). (D) Effect of 3-bp loop on the forward and reverse GRE (±SEM). The DNA sequence is shown for L = 5. koff is normalized to that measured on DNA without a hairpin loop.