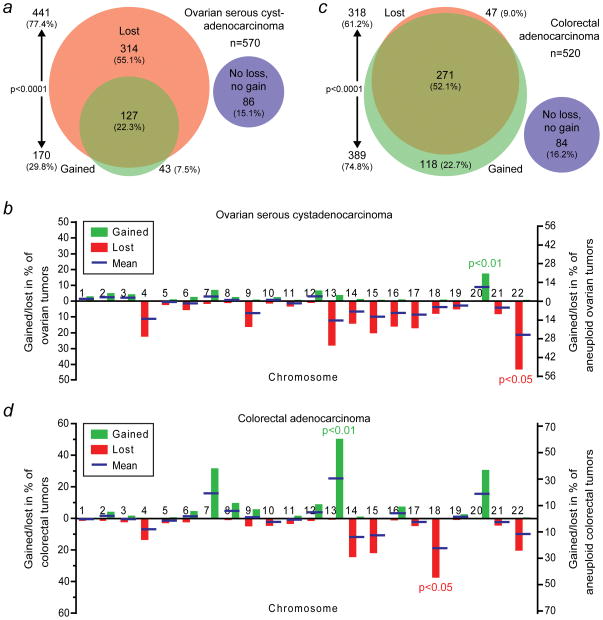
Figure 3.
Different types of human solid tumors have distinct whole-chromosome aneuploidy signatures. (a) Area-proportional Venn diagram as in Figure 1a. However, these data are derived from The Cancer Genome Atlas (TCGA) array-based comparative genomic hybridization (aCGH) data of 570 human ovarian serous cystadenocarcinomas. (b) Rates at which ovarian serous cystadenocarcinomas gain or lose individual whole-chromosomes. Blue bars indicate the means of the gain and loss rates. Chromosome 20 is gained significantly more frequently than any of the other chromosomes (p<0.01; two-sided Grubbs’ test), whereas chromosome 22 is significantly more frequently lost than any of the other chromosomes (p<0.05; two-sided Grubbs’ test). (c) Area-proportional Venn diagram of 520 human colorectal adenocarcinomas using aCHG-derived data from TCGA. The areas between (a) and (c) are not proportional. (d) Individual whole-chromosome gain and loss rates as in (b) but for colorectal adenocarcinomas. Chromosomes 13 and 18 are more frequently gained and lost, respectively, than the other chromosomes (p<0.01 and p<0.05, respectively; two-sided Grubbs’ tests).