Abstract
Ubiquitin C-terminal hydrolase-L1 (UCHL1), a neuron-specific de-ubiquitinating enzyme, is one of the most abundant proteins in the brain. We describe three siblings from a consanguineous union with a previously unreported early-onset progressive neurodegenerative syndrome featuring childhood onset blindness, cerebellar ataxia, nystagmus, dorsal column dysfuction, and spasticity with upper motor neuron dysfunction. Through homozygosity mapping of the affected individuals followed by whole-exome sequencing of the index case, we identified a previously undescribed homozygous missense mutation within the ubiquitin binding domain of UCHL1 (UCHL1GLU7ALA), shared by all affected subjects. As demonstrated by isothermal titration calorimetry, purified UCHL1GLU7ALA, compared with WT, exhibited at least sevenfold reduced affinity for ubiquitin. In vitro, the mutation led to a near complete loss of UCHL1 hydrolase activity. The GLU7ALA variant is predicted to interfere with the substrate binding by restricting the proper positioning of the substrate for tunneling underneath the cross-over loop spanning the catalytic cleft of UCHL1. This interference with substrate binding, combined with near complete loss of hydrolase activity, resulted in a >100-fold reduction in the efficiency of UCHL1GLU7ALA relative to WT. These findings demonstrate a broad requirement of UCHL1 in the maintenance of the nervous system.
Keywords: protein quality control, recessive inherited neurodegeneration
Neurodegenerative syndromes represent a diverse group of disorders characterized by progressive neurological decline, typically associated with an anatomical correlate. Although the clinical onset of most of the common neurodegenerative disorders, such as Parkinson or Alzheimer’s diseases, is during adulthood (1), some syndromes become manifested during childhood. Despite being exceedingly rare in the population, understanding the molecular basis of these early-onset neurodegenerative syndromes could allow a unique insight into mechanisms of central nervous system maintenance. To gain a biological understanding of such disorders, we focused on consanguineous kindreds in which the affected subjects presented to medical attention during childhood with neurological decline. In one such kindred, NG 1024 (Fig. 1A), which originated from Turkey, three of the six siblings, who were offspring of a first-cousin consanguineous union, were found to have vision loss during early childhood, followed by progressive neurological dysfunction of the pyramidal system, cerebellum, and spinal dorsal columns. To identify the genetic basis of this syndrome with apparent autosomal recessive mode of inheritance, we initially used whole genome genotyping to map the homozygous segments shared by affected family members, which presumably contained the disease-causing mutation. We then performed whole-exome capture and next-generation sequencing of the index case. Application of next-generation sequencing technologies, specifically exome sequencing, which allows for selective sequencing of all exons, has previously been proven to be the most efficient and cost-effective approach in the discovery of disease-causing variants in various Mendelian disorders (2–4), such as this family. Based on the results of the exome sequencing, we analyzed each variant located within the homozygous regions shared by all affected members. We identified a homozygous missense mutation affecting a glutamic acid residue within the ubiquitin binding domain of the ubiquitin C-terminal hydrolase L1 (UCHL1) gene (UCHL1GLU7ALA) that cosegregated with the phenotype.
Fig. 1.
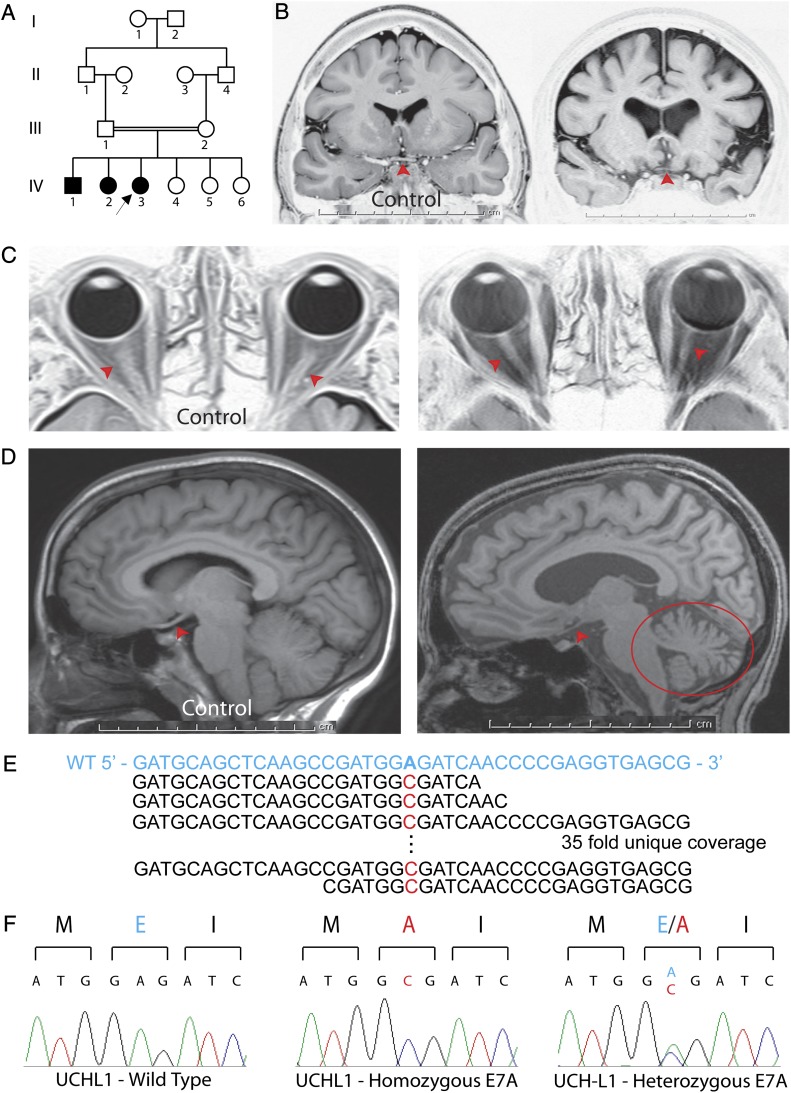
Identification of the GLU7ALA (E7A) mutation in UCHL1 in kindred NG 1024. (A) Simplified pedigree is shown; affected subjects are denoted by filled symbols (arrow indicates the index case). (B) Coronal MR images of NG 1024–3 (Right) and a control subject (Left) reveal diffuse cerebral and optic chiasmal atrophy (red arrowhead) in the index case. Cortical atrophy is indicated by cerebral volume loss with increase in the subarachnoid and intersulci spaces over the brain, which are filled with cerebrospinal fluid (black in these images). (C) Axial orbital MR images of a control subject (Left) and NG 1024–3 (Right) show optic nerve (red arrowhead) atrophy. (D) Sagittal MR images of a control subject (Left) and NG 1024–3 (Right) reveal cerebral cortical, cerebellar (red circle), and optic tract (red arrowheads) atrophy. (E) Exome sequencing reveals an A to C change. The WT sequence in blue is shown on top, with the mutant base in red below. There was 35-fold coverage, with all reads revealing the substitution. (F) Sanger sequencing confirms the mutation that results in glutamic acid (E, in blue) to alanine (A, in red) change. The unaffected parents are heterozygous for the variant (Right).
UCHL1 is one of the most abundant proteins in the brain, comprising 1–2% of the total soluble fraction (5). It is a neuron-specific de-ubiquitinating enzyme and plays an important role in ubiquitin turnover through its C-terminal hydrolytic activity. UCHL1 has also been suggested to be a ubiquitin ligase (6). Although UCHL1 has previously been implicated in the pathophysiology of neurodegenerative disorders including Parkinson (7) and Alzheimer’s (8) diseases, conclusive evidence that links UCHL1 dysfunction to neurodegeneration has been lacking. Here, based on the molecular genetics data that led to the identification of the UCHL1GLU7ALA mutation in the family under study, followed by the molecular studies that demonstrated the GLU7ALA mutation to nearly completely abolish UCHL1’s hydrolase activity, we conclude that compromised UCHL1 activity leads to a childhood-onset multisystem neurodegenerative syndrome. Our findings link loss of UCHL1 function with broad neurodegeneration and demonstrate its fundamental importance in the maintenance of the nervous system.
Results
Early-Onset Neurodegenerative Syndrome.
All three affected children were products of uncomplicated, term labors and reached early neurodevelopmental milestones normally. They began suffering from vision loss at around age 5 y, followed by slowly progressive neurological problems. After three decades, all had developed blindness, cerebellar ataxia with an inability to stand without assistance, nystagmus, and titubation (head tremor) (Table S1; SI Materials and Methods). Muscle strength was grossly normal in all subjects, with the male subject suffering from myotonia (muscles failing to relax after activity). There was decreased vibration and position sense due to dorsal column dysfunction. Deep tendon reflexes were increased throughout with spasticity, indicating upper motor neuron dysfunction. There were no extrapyramidal signs. Outside the nervous system, there was no apparent phenotype. The parents and remaining three siblings were healthy.
Further laboratory tests confirmed the findings on physical examination (Figs. S1 and S2). Consistent with the clinical finding of blindness, flash visual-evoked potentials revealed nearly absent response in both eyes but normal full-field electroretinograms. Nerve conduction velocities were normal, but myokymic activity was detected in all muscles tested during EMG (Fig. S1). Somatosensory-evoked potential (SSEP) studies suggested dorsal column dysfunction that was more significant in the lower extremities (Fig. S2).
In all three patients, MRI scans were remarkable for bilateral optic nerve and chiasm atrophy, with diffusion tensor imaging (DTI) studies revealing bilateral Wallerian degeneration of the optic radiations (Fig. 1 B–D; Table S2) (9, 10). The MRI scans were also remarkable for obvious cerebellar and mild cerebral atrophy that was more severe in the older siblings (Fig. 1 B and D). The finding of a rare phenotype recurring in a sibship from a consanguineous union suggests autosomal recessive transmission.
Whole-Exome Sequencing Identified a Missense Variant, GLU7ALA, in UCHL1.
We initially performed whole-genome genotyping of the three affected siblings using Illumina 610K SNP chips and confirmed the reported consanguinity. Slightly greater that what is to be expected for a consanguineous first-cousin union, inbreeding coefficients of the affected subjects ranged between 9.6% and 10.6%, and the number of the homozygous segments (>2.5 cM) in each patient was between 23 and 34, with the total size of the homozygous regions being between 241.3 and 358.7 million bp (MB) (324.08 and 382.09 cM, respectively). Among these homozygous segments, only four chromosomal regions, on chromosomes 4, 6, 8, and 11, were shared by all three affected siblings. These regions, which together comprised 27.16 MB, presumably contained the disease-causing mutation (Table S3). We next performed whole-exome sequencing of the index case using Nimblegen solid-phase array capture and the Illumina Genome Analyzer IIx instrument and focused on variants located within these shared regions (2–4, 11). A single lane of sequencing on Illumina’s Genome Analyzer IIX with single-read chemistry and a read length of 74 bases yielded ∼38 million reads. These data in turn achieved a high and uniform coverage across the targeted bases with a mean coverage of ∼52-fold. Nearly 99% and 97% of all of the targeted bases within the homozygosity intervals were read at least four and eight times, respectively. The mean sequence error rate was 0.97% (Table S4). The sensitivity and specificity for the detection of homozygous variation from the reference human genome were high (98.75% and 99.75%, respectively) as determined by comparison of sequencing data to the results of SNP genotyping as a reference.
Although a total of 141 homozygous variants were detected within these homozygous segments, only five of these variants were not previously identified in dbsnp (build 131) and 1,000 genomes project databases and among 2,400 exomes of European subjects sequenced at Yale (Table 1; Table S5), and two of the homozygous variants altered the encoded protein. The first of these variants, a proline to leucine substitution in ZNF259 (ZNF259PRO18LEU) (chromosome 11: 116,658,654 G > A), failed to segregate with the disease phenotype—the father and one of the unaffected siblings were homozygous for the variant. In contrast, the second variant, an A to C transversion on chromosome 4, position 41,259,013, cosegregated with the trait and was absent both from the Yale cohort and 948 Turkish control chromosomes (Table 1; Table S5). This single base substitution produced a missense mutation (GLU7ALA) at codon 7 of UCHL1 (NM_004181) (Fig. 1E), a position that is completely conserved among vertebrate orthologs and is predicted to be within the ubiquitin binding domain of the protein. The variant was confirmed to be homozygous in all three affected subjects, and neither parent nor unaffected siblings were homozygous for the variant by Sanger sequencing (Fig. 1F; Fig. S3). The cosegregation of this rare homozygous mutation with the neurodegeneration phenotype supported the hypothesis that it is the cause of this Mendelian trait.
Table 1.
Variants detected within shared homozygous segments
| Chromosome | Start* | End* | Variants | Previously unreported variants† | Previously unreported protein-altering variants | Previously unreported protein-altering variants that cosegregate with the phenotype |
| 4 | 38,187,084 | 49,061,848 | 66 | 3 | 1 | 1 |
| 6 | 44,575,446 | 53,593,569 | 49 | 1 | 0 | 0 |
| 8 | 39,869,633 | 43,791,691 | 9 | 0 | 0 | 0 |
| 11 | 114,223,577 | 117,570,851 | 17 | 1 | 1 | 0 |
*NCBI37/hg19.
†Variants that were not previously identified in dbsnp (build 131) and 1,000 genomes project databases and among 2,400 exomes of European subjects sequenced at Yale.
UCHL1GLU7ALA Is Predicted to Interfere with Substrate Binding.
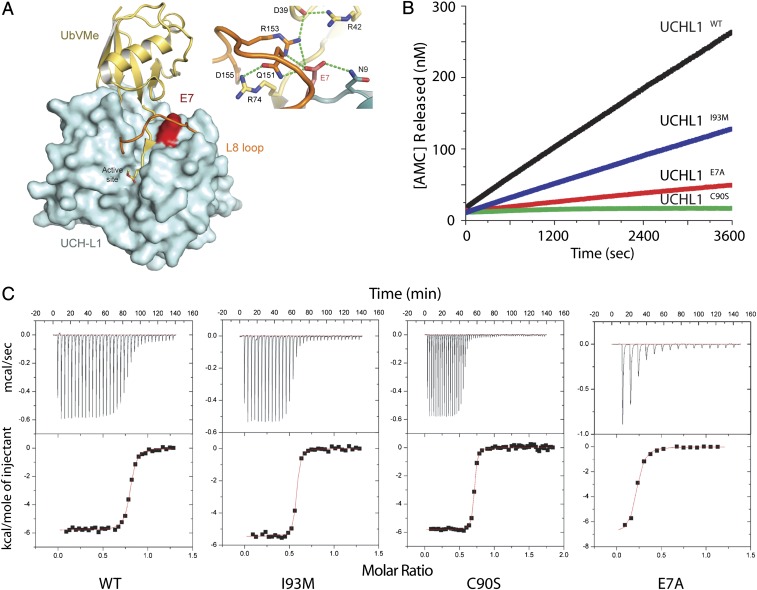
UCHL1 is a de-ubiquitinating enzyme, involved in the recycling of free ubiquitin and therefore cytoplasmic protein degradation through its C-terminal hydrolase enzymatic activity. To gain further insight into the role of UCHL1 in neurodegeneration, we examined the structural implications and functional consequences of the GLU7ALA mutation. The tertiary structure of UCHL1 resembles that of the papain family (12) and is characterized by the presence of a cross-over loop, termed L8, that spans and thereby restricts access to the catalytic cleft that consists of the histidine-cysteine-aspartic acid triad (12, 13) (Fig. 2A). In UCHL1, the L8 loop is short, thus requiring the ubiquitin substrate to tunnel underneath it to achieve proteolysis; residue E7 is located directly at the threshold of this tunnel (12, 13). In the complex of UCHL1 and the ubiquitin substrate mimic, ubiquitin vinyl methyl ester (UbVMe) (13), GLU7 is involved in a hydrogen-bonding network that interacts both with the ubiquitin substrate mimic and the L8 loop (Fig. 2A). The GLU7ALA variant is therefore predicted to restrict the proper positioning of the substrate for tunneling underneath the L8 loop, without conferring any other significant effects on the overall tertiary structure, e.g., exhibiting a circular dichroism (CD) spectrum virtually identical to WT.
Fig. 2.
Structural and functional studies of UCHL1 carrying the GLU7ALA mutation. (A) Structural mapping of GLU7ALA (E7A) onto the crystal structure of UCHL1 in complex with the ubiquitin substrate mimic, UbVMe (13) (PDB ID: 3IFW). UCHL1 is colored cyan, with the L8 loop colored orange. UbVMe is colored yellow. Residue E7 is colored red. (Inset) Close-up of the hydrogen-bonding network at the threshold of the substrate tunnel. Panel made using the program Pymol (www.pymol.org). (B) Comparison of the enzymatic activity of WT UCHL1 and the variants I93M, C90S, and E7A. Real-time release of fluorescent AMC is shown for WT-UCHL1 (black), I93M (blue), E7A (red), and C90S (green). (C) Binding isotherms of the titration of WT-UCHL1 and respective mutants with ubiquitin. Binding of ubiquitin (500 μM) to corresponding proteins (50 μM) is shown. (Upper) Raw data. (Lower) Integrated heat data as enthalpy as a function of molar ratio of ligand to protein. The solid line in the lower panel represents the best curve fit to the data by using a one-site binding model.
UCHL1GLU7ALA Results in Reduced Binding Affinity for Ubiquitin and Severely Reduced Hydrolytic Activity.
We next analyzed the effects of the GLU7ALA mutation on ubiquitin binding and C-terminal hydrolase enzymatic activity. In Escherichia coli, we expressed and purified WT UCHL1 (UCHL1WT) and mutants UCHL1GLU7ALA, UCHL1ILE93MET, and UCHL1CYS90SER (a cysteine to serine substitution in position 90), which carries a mutation in the active cleft consisting of the abovementioned triad and was previously shown to interfere with the catalytic activity (12–14). First, the binding of ubiquitin to these purified proteins was measured using isothermal titration calorimetry, from which the Kd values for ubiquitin binding to these proteins were calculated (Fig. 2C; Table S6). For the WT protein, the Kd was 85 ± 31 nM, and similar Kds were determined for the CYS90SER and ILE93MET versions. By contrast, GLU7ALA had a much higher Kd, indicating at least sevenfold decreased ubiquitin binding (Table 2).
Table 2.
Binding and thermodynamic parameters for UCHL1WT, UCHL1GLU7ALA, UCHL1CYS90SER, and UCHL1ILE93MET
| Parameters | UCHL1WT | UCHL1GLU7ALA | UCHL1CYS90SER | UCHL1ILE93MET |
| kcat (s−1) | 0.188 ± 0.006 | 0.017 ± 0.005 | ND | 0.084 ± 0.003 |
| Kd (nM)* | 85 ± 31 | 646 ± 288 | 32 ± 6 | 29 ± 5 |
| kcat/Kd (nM−1⋅s−1) | 2.2 × 10−3 | 2.6 × 10−5 | ND | 2.8 × 10−3 |
Values shown are the average of three different experiments ± SD. kcat, catalytic constant; Kd, dissociation constant; ND, not detectable.
*Kd values for all proteins were determined by isothermal titration calorimetry.
Next, the enzymatic activity of the purified enzymes was examined. In terms of its effects on catalytic activity, in an assay with ubiquitin-7-amido-4-methycoumarin (ubiquitin-AMC) as substrate where ubiquitin-AMC cleavage was continuously monitored and measured on a fluorescence spectrometer, UCHL1GLU7ALA exhibited <10% hydrolase activity compared with WT, whereas UCHL1ILE93MET exhibited the previously reported ∼50% activity (7); as expected, UCHL1CYS90SER exhibited no hydrolase activity (Fig. 2B). Overall, the efficiency of UCHL1GLU7ALA, as measured by kcat/Kd, was 100-fold reduced relative to WT and UCHL1ILE93MET (Table 2).
It has previously been suggested that UCHL1 may exhibit novel dimerization-dependent ubiquitin ligase activity in vitro (6). In an attempt to determine the effects of the GLU7ALA variant on the suggested ubiquitin ligase activity of UCHL1, we assayed and tested the enzymatic activity as previously described (6). Neither UCHL1WT nor UCHL1GLU7ALA exhibited ubiquitin ligase activity and did not ubiquitinate α-synuclein. Further studies are needed to characterize the complete role of UCHL1 in proteasomal protein degradation.
Discussion
Our molecular and biochemical data link the homozygous UCHL1GLU7ALA mutation, which severely compromises ubiquitin binding and hydrolase activity, with an early-onset progressive degenerative syndrome affecting multiple pathways within the nervous system including the optic system, cerebral cortex, cerebellum, and spinal cord. In addition, myokymia was observed in affected subjects, consistent with UCHL1’s role in the neuromuscular junction (15). The ataxia and muscular phenotypes in individuals with the homozygous UCHL1GLU7ALA mutation are consistent with the phenotypes of the gracile axonal dystrophy (gad) mouse, harboring a spontaneous in-frame deletion in Uchl1 that results in a truncated protein, as well as those of the Uchl1 KO mice that lack the entire Uchl1 protein (15–17). The gad mouse has been reported to suffer from a progressive neurological phenotype characterized by development of tremor and sensory ataxia at around 3 mo of age postnatally, followed by motor ataxia and ultimately leading to mortality. Neuropathological findings included nerve fiber loss with astrocytic proliferation and considerable axonal swellings in the gracile fascicles of the spinal cord along with axonal degeneration and formation of spheroid bodies in the nerve terminals (16, 18, 19). Similarly, recently generated Uchl1 KO mice have been shown to suffer from markedly impaired synaptic transmission at the neuromuscular junction accompanied by structural defects such as loss of synaptic vesicles and accumulation of tubulovesicular structures at the presynaptic nerve terminals in addition to denervation of the muscles (15). The phenotypes of both of these Uchl1 mouse models closely resemble that of our patients, with the exception of lack of optic nerve degeneration in the Uchl1 KO mice, examined at about 4 mo of age. Further analyses of these models at later stages will be needed.
In 1998, a heterozygous missense variant (ILE93MET) in UCHL1 was described in a German family with Parkinson disease and suggested as the cause of autosomal dominant Parkinson disease type 5 (PARK5, OMIM 613643) (7). The UCHL1ILE93MET variant results in a 50% reduction in hydrolytic activity; therefore, the heterozygous affected subjects are expected to have 75% of normal activity. Subsequent studies failed to replicate the ILE93MET variant in familial and sporadic forms of Parkinson disease and identified only one other common UCHL1 variant, a serine to tyrosine (SER18TYR) polymorphism (20, 21). Although a protective effect for the UCHL1SER18TYR variant against Parkinson disease was initially suggested, several subsequent association studies yielded conflicting results (22–24). In the family presented here, neither the patients homozygous for the UCHL1GLU7ALA variant nor their heterozygous parents or siblings exhibited Parkinsonian features on neurological examination. This finding might be due to phenotypic heterogeneity associated with the different UCHL1 variants (heterozygous ILE93MET mutation vs. homozygous GLU7ALA mutation).
In addition to these genetic data, UCHL1’s expression has been shown to be down-regulated in the brains of patients with both Parkinson and Alzheimer’s diseases (25). UCHL1 has been detected in neurofibrillary tangles in idiopathic Alzheimer’s disease patients, with the levels of soluble UCHL1 being inversely proportional to the number of tangles (25). Finally, it has been shown that UCHL1 activity is required for normal synaptic function and may improve the retention of memory in a mouse model of Alzheimer’s disease (8). Collectively, these studies suggest an association between UCHL1 dysfunction and neurodegeneration, which is strengthened by our discovery of a mutation that almost completely abolishes its hydrolytic activity and interferes with ubiquitin binding in a family with a neurodegenerative syndrome. Our findings further demonstrate the fundamental importance of protein degradation and the ubiquitin pathway in proper functioning of the nervous system. This observation is in agreement with the recent identification of Ubiquilin 2 (UBQLN2) mutations in another neurodegenerative syndrome, familial amyotrophic lateral sclerosis (ALS) (26). Loss of UBQLN2 function leads to impaired protein degradation and accumulation of UBQLN2-containing protein aggregates in spinal motor neurons of ALS patients. Interestingly, UBQLN2 accumulated in spinal cord inclusion bodies irrespective of the patients carrying UBQLN2 mutations. A similar pathology is observed in the hippocampi of patients with ALS and dementia, again with or without UBQLN2 mutations (26), implying that impairment of ubiquitin-dependent proteolysis has pleiotropic effects leading to broad degeneration of the nervous system.
Our results strengthen the link between ubiquitin-dependent proteolysis and nervous system maintenance and demonstrate that loss of UCHL1 activity results in a childhood-onset neurodegenerative syndrome. The observed phenotype is progressive, leading to diffuse cerebral and cerebellar atrophy with aging. Identifying the normal targets of UCHL1 and their biochemical functions will provide insight into the maintenance of nervous system integrity.
Materials and Methods
Human Subjects.
The study protocol was approved by the Yale Human Investigation Committee (protocol no. 0908005592). Institutional review board approval for genetic studies, along with written consent from all study subjects, was obtained at the participating institution.
EMG and SSEP Studies.
Intramuscular needle EMG studies were performed in all three patients testing deltoid and gastrocnemius muscles using conventional techniques. Conduction velocities and response amplitudes were measured for both upper and lower peripheric motor (median, ulnar, peroneal and tibial) and sensory (median, ulnar and sural) nerves. For SSEP studies, scalp evoked potential responses were recorded at 100 ms following right tibial and median nerve stimulation in all three patients.
MRI.
All MRI examinations were performed in a 3T scanner (Trio, Siemens) using 8-channels head coil. The DTI datasets were obtained and analyzed as described before (9). Briefly, high resolution cranial MRIs were performed using the following sequences: sagittal TSE T2, axial SE T1, TSE T2, FLAIR and diffusion, coronal TSE T2, high resolution 3D TurboFlair T2 and reconstructions, sagittal 3D TurboFlash T1 and reconstructions, sagittal 3D SPACE T2 and reconstructions, coronal optic nerve focused TSE - STIR, SE T1, DTI- FT, axial GRE T2.
Illumina Genotyping.
Whole-genome genotyping of the samples was performed on the Illumina Platform with Illumina Human 610K Quad Beadchips using the manufacturer's protocol (Illumina) and as previously described (2, 3, 11).
Whole-Exome Capture and Sequencing.
Genomic DNA sample was captured on a NimbleGen 2.1M human exome array version 1.0 (Roche Nimblegen, Inc.) with modifications to the manufacturer's protocol (2–4, 11). Briefly, after quality assessment, the genomic DNA was sheared by sonication, the ends of fragments were repaired, adaptors were ligated, and appropriately sized fragments were selected using agarose gel electrophoresis. Precapture ligation mediated PCR was performed, purified DNA was hybridized to the array, and after multiple washes, the DNA was eluted and amplified by postcapture mediated PCR. The pre- and post- capture libraries were compared by quantitative PCR for the determination of the relative fold enrichment of the targeted sequences. Single-read cluster generation was performed on the Cluster Station (Illumina) and the captured, purified, and clonally amplified library was then sequenced on Genome Analyzer IIx. One lane of single-read sequencing at a read length of 74 bases was performed on Genome Analyzer IIx following the manufacturer’s protocol.
Analysis of the Whole-Exome Sequencing Data.
The sequence reads obtained were aligned to the human genome (hg18) using Maq (27) and BWA (28) software as previously described (4). Perl scripts were used to calculate the percentage alignment of the reads to the reference genome and the targeted exome. Similarly, perl scripts were used for the detection of mismatch frequencies and error positions. SAMtools (29) was used for the detection of single nucleotide variations on the reads aligned with Maq. The indels were detected on the reads aligned with BWA for its ability to allow for gaps during the alignment. Shared homozygous segments of the affected individuals were detected using Plink software version 1.07 (30), and the variants were filtered for shared homozygosity. The variants were annotated for novelty with comparison with dbSNP (build 131), 1000 Genomes database (August 4, 2010 release) and previous exome sequencing experiments performed by our human genomics groups.
Sanger Sequencing.
The exons and exon-intron boundaries of UCHL1 were determined using the UCSC Genome Browser (http://genome.ucsc.edu), unique primers were designed using Sequencher 4.8 (Gene Codes) and were synthesized by Invitrogen. The fragments were amplified using standard PCR techniques and sequenced on ABI's 9800 Fast Thermocyclers (Applied Biosystems Inc.).
CD Spectroscopy.
To determine CD spectra, stock solutions of UCHL1WT, UCHL1GLU7ALA, UCHL1CYS90SER, and UCHLILE93MET mutants were diluted to a final concentration of 10 μM with 50 mM Tris-HCl, pH 7.4 buffer. CD spectra were recorded by using an Applied Photophysics spectropolarimeter in the wavelength scan mode. Typically, data were obtained as an average of three scans in the wavelength region of 280–195 nm by using a quartz cuvette of 0.1-cm path.
Isothermal Titration Calorimetry.
Isothermal titration calorimetry (ITC) was carried out by using a VP-ITC Microcal calorimeter (Microcal) at 24 °C. All protein samples were extensively dialyzed against 50 mM Tris-HCl, pH 7.6. Titrations consisted of 5-μL injections of ubiquitin (Boston Biochem) into the sample cell containing the respective UCHL1, at time intervals of 4 min to ensure each peak returned to baseline. Each UCHL1 sample was followed by a background titration of an equal volume of ubiquitin being titrated into a sample cell containing buffer only, to account for the heat of ubiquitin dilution, which was subtracted from the ubiquitin-UCHL1 data. All data were analyzed by using the program Origin, version 7.0, included with the system. The data were fitted with a one-site binding model. Binding constants and thermodynamic parameters of three experiments (average ± SD) are given.
Enzymatic Activity Assay.
Wild type UCHL1 and the GLU7ALA, ILE93MET, and CYS90SER mutants were expressed in E.coli and purified as previously described (14). Purified proteins were diluted into reaction buffer (50 mM Tris-HCl, pH 7.4, 1 mM DTT, and 1 mM EDTA) in a 200 µl fluorescence cuvette to a final concentration of 3 nM. Ubiquitin-AMC was added to the reaction mixture to yield a final concentration of 2500 nM to initiate the enzymatic reaction. AMC cleavage was monitored at 25 °C on a PTI QuantaMaster fluorescence spectrometer with excitation at 380 nm and emission at 465 nm.
Supplementary Material
Acknowledgments
We thank the patients and families who contributed to this study. This work was supported by the Yale Program on Neurogenetics, National Institutes of Health (NIH) Grants RC2 NS070477 (to M.G.) and UL1 RR024139NIH (to S.M.), Yale Center for Mendelian Disorders Grant U54HG006504 (to R.P.L., M.G., and S.M.), and the Gregory M. Kiez and Mehmet Kutman Foundation. R.P.L. and A.L.H. are Investigators of the Howard Hughes Medical Institute. SNP genotyping was supported in part by NIH Neuroscience Microarray Consortium Award U24 NS051869-02S1 (to S.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222732110/-/DCSupplemental.
References
- 1.Ballard C, et al. Alzheimer’s disease. Lancet. 2011;377(9770):1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- 2.Barak T, et al. Recessive LAMC3 mutations cause malformations of occipital cortical development. Nat Genet. 2011;43(6):590–594. doi: 10.1038/ng.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilgüvar K, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467(7312):207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi M, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106(45):19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson KD, et al. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science. 1989;246(4930):670–673. doi: 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111(2):209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 7.Leroy E, et al. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395(6701):451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 8.Gong B, et al. Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell. 2006;126(4):775–788. doi: 10.1016/j.cell.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 9.Dinçer A, et al. Diffusion tensor imaging of Guillain-Mollaret triangle in patients with hypertrophic olivary degeneration. J Neuroimaging. 2011;21(2):145–151. doi: 10.1111/j.1552-6569.2009.00461.x. [DOI] [PubMed] [Google Scholar]
- 10.Salmela MB, Cauley KA, Nickerson JP, Koski CJ, Filippi CG. Magnetic resonance diffusion tensor imaging (MRDTI) and tractography in children with septo-optic dysplasia. Pediatr Radiol. 2010;40(5):708–713. doi: 10.1007/s00247-009-1478-0. [DOI] [PubMed] [Google Scholar]
- 11.Bakircioglu M, et al. The essential role of centrosomal NDE1 in human cerebral cortex neurogenesis. Am J Hum Genet. 2011;88(5):523–535. doi: 10.1016/j.ajhg.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das C, et al. Structural basis for conformational plasticity of the Parkinson’s disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci USA. 2006;103(12):4675–4680. doi: 10.1073/pnas.0510403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boudreaux DA, Maiti TK, Davies CW, Das C. Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCHL1 into productive conformation. Proc Natl Acad Sci USA. 2010;107(20):9117–9122. doi: 10.1073/pnas.0910870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen CN, Price JS, Wilkinson KD. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: Identification of two active site residues. Biochemistry. 1996;35(21):6735–6744. doi: 10.1021/bi960099f. [DOI] [PubMed] [Google Scholar]
- 15.Chen F, Sugiura Y, Myers KG, Liu Y, Lin W. Ubiquitin carboxyl-terminal hydrolase L1 is required for maintaining the structure and function of the neuromuscular junction. Proc Natl Acad Sci USA. 2010;107(4):1636–1641. doi: 10.1073/pnas.0911516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saigoh K, et al. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet. 1999;23(1):47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- 17.Yamazaki K, et al. Gracile axonal dystrophy (GAD), a new neurological mutant in the mouse. Proc Soc Exp Biol Med. 1988;187(2):209–215. doi: 10.3181/00379727-187-42656. [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi T, Mukoyama M, Yamazaki K, Moriya H. Axonal degeneration of ascending sensory neurons in gracile axonal dystrophy mutant mouse. Acta Neuropathol. 1990;80(2):145–151. doi: 10.1007/BF00308917. [DOI] [PubMed] [Google Scholar]
- 19.Mukoyama M, Yamazaki K, Kikuchi T, Tomita T. Neuropathology of gracile axonal dystrophy (GAD) mouse. An animal model of central distal axonopathy in primary sensory neurons. Acta Neuropathol. 1989;79(3):294–299. doi: 10.1007/BF00294664. [DOI] [PubMed] [Google Scholar]
- 20.Lincoln S, et al. Low frequency of pathogenic mutations in the ubiquitin carboxy-terminal hydrolase gene in familial Parkinson’s disease. Neuroreport. 1999;10(2):427–429. doi: 10.1097/00001756-199902050-00040. [DOI] [PubMed] [Google Scholar]
- 21.Wintermeyer P, et al. Mutation analysis and association studies of the UCHL1 gene in German Parkinson’s disease patients. Neuroreport. 2000;11(10):2079–2082. doi: 10.1097/00001756-200007140-00004. [DOI] [PubMed] [Google Scholar]
- 22.Healy DG, et al. UCHL-1 is not a Parkinson’s disease susceptibility gene. Ann Neurol. 2006;59(4):627–633. doi: 10.1002/ana.20757. [DOI] [PubMed] [Google Scholar]
- 23.Maraganore DM, et al. UCHL1 Global Genetics Consortium UCHL1 is a Parkinson’s disease susceptibility gene. Ann Neurol. 2004;55(4):512–521. doi: 10.1002/ana.20017. [DOI] [PubMed] [Google Scholar]
- 24.Ragland M, Hutter C, Zabetian C, Edwards K. Association between the ubiquitin carboxyl-terminal esterase L1 gene (UCHL1) S18Y variant and Parkinson’s Disease: A HuGE review and meta-analysis. Am J Epidemiol. 2009;170(11):1344–1357. doi: 10.1093/aje/kwp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi J, et al. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004;279(13):13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 26.Deng HX, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477(7363):211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18(11):1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.