Abstract
Germ cell fate decisions are poorly understood, despite their central role in reproduction. One fundamental question has been whether germ cells are regulated to enter the meiotic cell cycle (i.e., mitosis–meiosis decision) and to be sperm or oocyte (i.e., sperm–oocyte decision) through one or two cell fate choices. If a single decision is used, a male-specific or female-specific meiotic entry would lead necessarily toward spermatogenesis or oogenesis, respectively. If two distinct decisions are used, meiotic entry should be separable from specification as sperm or oocyte. Here, we investigate the relationship of these two decisions with tools uniquely available in the nematode Caenorhabditis elegans. Specifically, we used a temperature-sensitive Notch allele to drive germ-line stem cells into the meiotic cell cycle, followed by chemical inhibition of the Ras/ERK pathway to reprogram the sperm–oocyte decision. We found that germ cells already in meiotic prophase can nonetheless be sexually transformed from a spermatogenic to an oogenic fate. This finding cleanly uncouples the mitosis–meiosis decision from the sperm–oocyte decision. In addition, we show that chemical reprogramming occurs in a germ-line region where germ cells normally transition from the mitotic to the meiotic cell cycle and that it dramatically changes the abundance of key sperm–oocyte fate regulators in meiotic germ cells. We conclude that the C. elegans mitosis–meiosis and sperm–oocyte decisions are separable regulatory events and suggest that this fundamental conclusion will hold true for germ cells throughout the animal kingdom.
Germ cells make two major cell fate choices. One is a cell cycle decision and one is a sexual fate decision. During the course of their development, germ cells are regulated to transition from the mitotic to the meiotic cell cycle, and they are regulated to produce sperm in males or oocytes in females. A fundamental question in the germ cell field has been whether the mitosis–meiosis and sperm–oocyte decisions represent one or two regulatory events (1). If a single decision, germ cells might decide between male-specific meiotic entry leading to spermatogenesis or female-specific meiotic entry leading to oogenesis. If two distinct decisions, germ cells would be regulated to enter the meiotic cell cycle in a way that is separable from their sexual fate decision. Major progress has been made in understanding regulation of the mitosis–meiosis decision (2), but the sperm–oocyte decision has been less tractable in most organisms. Here we investigate the relationship between the mitosis–meiosis and sperm–oocyte decisions in the nematode Caenorhabditis elegans, in which molecular regulators of the two decisions can be manipulated independently.
C. elegans adults exist as XO males or self-fertile XX hermaphrodites. Males produce sperm continuously; hermaphrodites make oocytes continuously in adults, after generating a limited number of sperm as larvae. In both sexes, the adult germ-line tissue is organized linearly along its distal to proximal axis: at the distal end, germ cells in the mitotic cell cycle occupy the “mitotic zone” (MZ); more proximally, germ cells enter the meiotic cell cycle and progress through meiotic prophase; germ cells terminally differentiate as sperm or oocytes at the proximal end (Fig. 1A). Germ cells move from distal to proximal as they progressively mature.
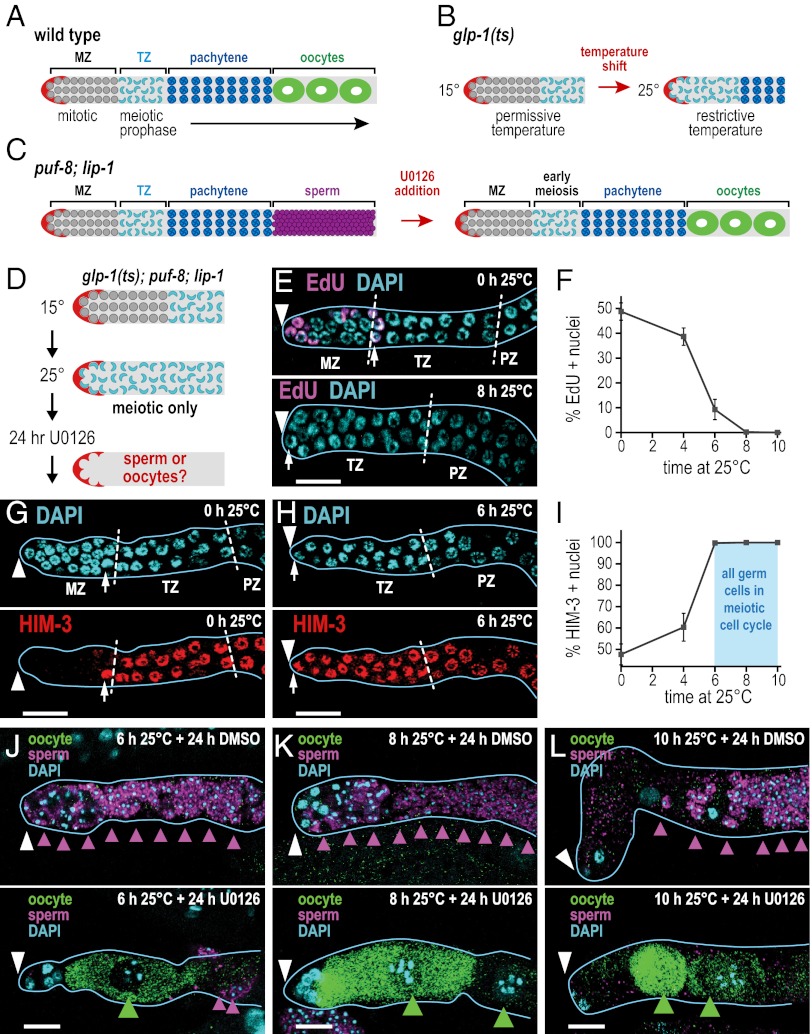
Fig. 1.
The mitosis–meiosis and sperm–oocyte cell fate decisions are separable. (A) Organization of WT C. elegans adult germ line. The stem cell niche (red) resides at the distal end; mitotic germ cells (gray) occupy the MZ; germ cells then enter the meiotic cell cycle and progress through early meiotic prophase (aqua-colored crescents) in the TZ and through pachytene (blue) in the pachytene zone (PZ). XX adult hermaphrodites make only oocytes (green). (B) Distal end of glp-1(ts) adult gonad. Germ cells are in the mitotic cell cycle at permissive temperature (15 °C) but enter the meiotic cell cycle when shifted to restrictive temperature (25 °C). (C) XX puf-8;lip-1 mutants make only sperm (purple), but treatment with the MEK inhibitor U0126 induces functional oocytes. (D) Experimental design to test separation of mitosis–meiosis and sperm–oocyte decisions. glp-1(ts);puf-8;lip-1 triple mutants are shifted to restrictive temperature (25 °C) to drive all germ cells into meiotic prophase (aqua-colored crescents); then U0126 or a DMSO solvent control is added, and germ lines are scored 24 h later for sperm or oocytes. (E, G, H, and J–L) Extruded gonads outlined in light blue; white arrowhead marks distal end; white dashed lines mark zone boundaries; small white arrow marks most distal meiotic prophase crescent. (Scale bars, 10 μm.) (E) At 15 °C (Upper), EdU labeling reveals S-phase nuclei in MZ, and DAPI shows meiotic prophase crescents, beginning at row 10. After 8 h at 25 °C, EdU incorporation ceases and crescent-shaped nuclei extend to distal end (Lower). (F) EdU incorporation as a function of time at 25 °C. Few S-phase cells remain at 6 h, and none remain at 8 h. Plot shows means ± SEM. (G) At 15 °C, the MZ lacks HIM-3 and crescents begin at row 10. (H) At 6 h after the shift to 25 °C, the MZ has been replaced by HIM-3+ meiotic cells that form crescents. (I) Percentage of HIM-3+ cells present in the distal germ line (i.e., MZ plus distal TZ) as a function of time shifted to 25 °C; light blue marks when all germ cells have entered the meiotic cell cycle. Points show means ± SEM. (J–L) Gamete sex can be reprogrammed from sperm to oocyte in meiotic germ cells. Sperm were visualized with sperm-specific marker SP56 (purple arrowheads); oocytes were seen with oocyte-specific marker OMA-2 (green arrowheads). % oo+, percentage of oocyte-positive animals. (J) Animals shifted to 25 °C for 6 h (U0126, 42% oocyte-positive animals, n = 12; DMSO, 0% oocyte-positive animals, n = 17); (K) animals shifted to 25 °C for 8 h (U0126, 52% oocyte-positive animals, n = 27 germ lines; DMSO, 0% oocyte-positive animals, n = 20); and (L) animals shifted to 25 °C for 10 h (U0126, 44% oocyte-positive animals, n = 32; DMSO, 4% oocyte-positive animals, n = 27 germ lines).
Regulators of the mitosis–meiosis decision have been identified and can be manipulated in C. elegans (3). The stem cell niche at the distal end (Fig. 1A, red) employs Notch signaling to maintain germ cells in the mitotic cell cycle and prevent meiotic entry. Without the C. elegans Notch receptor GLP-1 (Germ Line Proliferation-1), all mitotic germ cells enter the meiotic cell cycle and differentiate. A glp-1 ts (temperature-sensitive) mutant maintains the MZ at permissive temperature (15 °C), but, at the restrictive temperature (25 °C), all germ cells (including germ-line stem cells) enter the meiotic cell cycle (Fig. 1B). Therefore, glp-1(ts) mutants provide a powerful tool for manipulating the mitosis–meiosis decision in adults.
Regulators of the sperm–oocyte decision have also been identified and can be manipulated in C. elegans (3, 4). C. elegans germ cell sex relies on somatic signaling plus germ cell-specific sperm–oocyte fate regulators that respond to the somatic signals (3, 5). By manipulating sperm–oocyte fate regulators with temperature-sensitive mutants, RNA-mediated interference, or small molecule intervention, adults making sperm can be transformed to make oocytes without affecting somatic sex (e.g., refs. 4, 6, 7). Such a transformation from sperm to oocyte production can be induced in WT XO adult males or in XX adult hermaphrodites with an aberrantly masculinized germ line. Adults making oocytes can also be transformed to make sperm, again without affecting somatic sex (8). Germ-line sexual transformation does not appear to convert mature sperm into oocytes or vice versa, but instead switches the adult tissue from production of a gamete of one sex (e.g., sperm) into production of the other (e.g., oocyte).
One sperm–oocyte fate regulator is the C. elegans homologue of ERK/MAPK, called MPK-1 (9). We previously found that chemical inhibitors of Ras/ERK signaling can reprogram adults from sperm to oocyte production when applied in a puf-8;lip-1 sensitized mutant background (4) (Fig. 1C). XX puf-8;lip-1 adult germ lines make sperm instead of oocytes, likely because of the dual loss of the puf-8 oocyte fate regulator (10) and the lip-1 dual specificity phosphatase, which leads to hyperactivation of the MPK-1/ERK sperm fate regulator (9, 11). Importantly, this chemically induced oogenesis generates functional oocytes that support embryogenesis (4). Treatment with Ras/ERK inhibitors therefore provides a method to manipulate the sperm–oocyte decision quickly (mature oocytes are seen 24 h after treatment) and independently of a temperature shift.
In this study, we manipulate the mitosis–meiosis and sperm–oocyte decisions independently and find that they can be uncoupled: germ cells in meiotic prophase can be sexually transformed from a spermatogenic to an oogenic cell fate. We also find that the sperm-to-oocyte fate chemical reprogramming occurs in a region in the distal germ line where cells transition from the mitotic to the meiotic cell cycle, and that it is accompanied by dramatic changes in key sperm–oocyte fate regulators. Together these results provide compelling evidence that the mitosis–meiosis and sperm–oocyte decisions are separable regulatory events.
Results and Discussion
Mitosis–Meiosis and Sperm–Oocyte Decisions Are Separable.
We used a simple experimental design to ask if the mitosis–meiosis and sperm–oocyte decisions are separable regulatory events (Fig. 1D). We first generated glp-1(ts);puf-8;lip-1 triple mutants, a strain in which glp-1(ts) permitted temperature manipulation of the mitosis–meiosis decision and puf-8;lip-1 permitted chemical manipulation of the sperm–oocyte decision. We then asked how long after being shifted to restrictive temperature (25 °C) germ cells entered the meiotic cell cycle. Specifically, we assayed S-phase with incorporation of the thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU) (12) and meiotic entry with staining of chromosomal HIM-3, a synaptonemal complex protein and established marker of cells in the meiotic cell cycle (13). In both assays, DAPI staining was also used to monitor nuclear morphology diagnostic of various stages of meiotic prophase. We found that EdU incorporation ceased within 8 h after the shift to 25 °C (Fig. 1 E and F), suggesting that mitotic and meiotic S-phase were complete by that time. We also found that chromosomal HIM-3 was present in all germ cells within 6 h after the shift (Fig. 1 G–I), suggesting that meiotic entry was complete. By nuclear morphology, most germ cells had entered early meiotic prophase 6 h after the shift, and all had done so 8 h after the shift (Fig. S1). Therefore, a shift of glp-1(ts);puf-8;lip-1 triple mutants to 25 °C drives most germ cells into meiotic prophase by 6 h, and all germ cells are in meiotic prophase by 8 h.
To ask if we could reprogram meiotic prophase germ cells from sperm to oocyte production, we shifted the glp-1(ts);puf-8;lip-1 triple mutants to restrictive temperature for 6, 8, or 10 h and then applied the MEK kinase inhibitor U0126 and scored gametes after 24 h (Fig. 1D). Production of mature sperm or oocytes was scored by using gamete-specific molecular markers (sperm, SP56; oocyte, OMA-2), cellular morphology (3), and embryo generation. None of the DMSO solvent-treated control germ lines made oocytes (Fig. 1 J–L, Upper), but U0126-treated germ lines were reprogrammed from sperm to oocyte production (Fig. 1 J–L, Lower). Moreover, the reprogrammed germ lines made functional oocytes, as assayed by embryo production. One might like to ask if meiotic germ cells could similarly be forced from oocyte to sperm generation, but this opposite transformation cannot yet be exerted chemically. We conclude that the sperm–oocyte fate decision can be reprogrammed after entry into meiotic prophase, a result that unambiguously shows that the mitosis–meiosis and sperm–oocyte decisions are separable regulatory events. A corollary to this conclusion is that other sex-specific features of germ cells (e.g., mitotic cell cycle lengths, meiotic progression rates) may also be separable from the sperm–oocyte fate decision.
Sperm–Oocyte Fate Reprogramming Maps to Transition from Mitotic to Meiotic Cell Cycle.
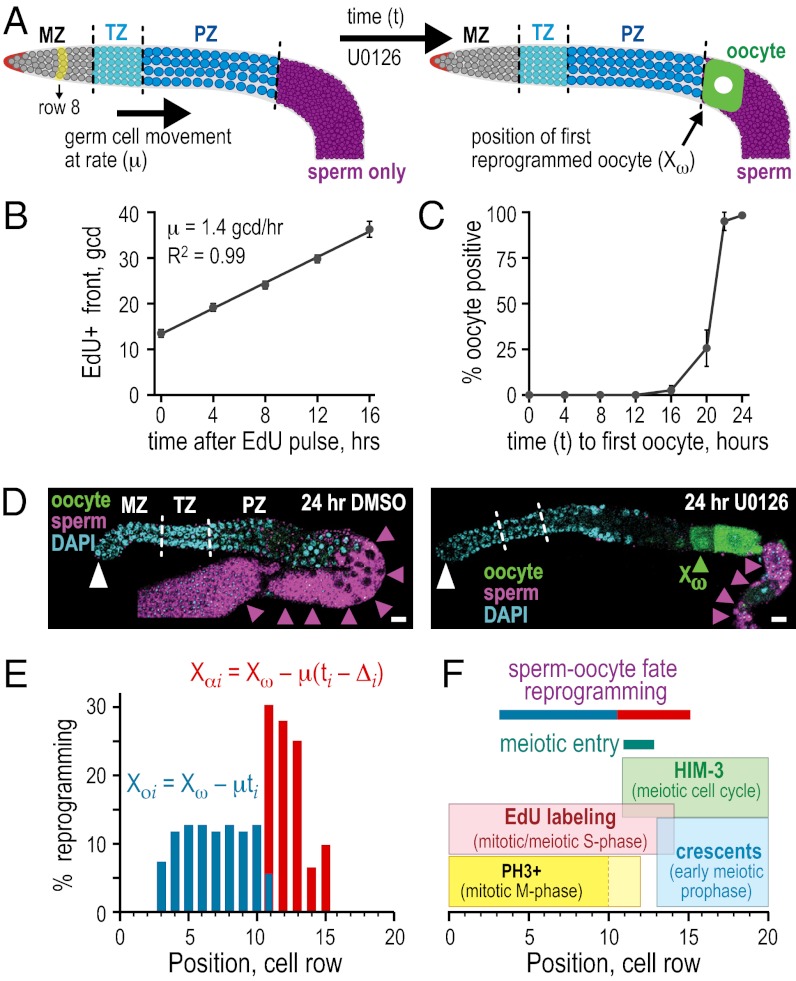
By using the puf-8;lip-1 double mutant on its own [without the glp-1(ts) mutation], we generated a map of the anatomical region where germ cells undergo the process of sperm–oocyte fate reprogramming. We subsequently mapped the site of meiotic entry in this same mutant for comparison. By convention, germ cell position is measured along the gonadal distal to proximal axis in number of germ cell diameters (gcds) from the distal end; because several germ cells reside at each position, each position is called a row for brevity (see Fig. 3A). To deduce the site of reprogramming, we measured several values, all in U0126-treated puf-8;lip-1 adult germ lines. The rate of germ cell movement, μ, was 1.4 gcds/h (Fig. 2B and Fig. S2); the position at which the first grossly differentiated oocyte appeared, Xω, was 34.4 gcds ± 1.8 (n = 29); and the time elapsed from U0126 addition to appearance of the first induced oocyte, t, spanned 16 to 22 h (Fig. 2 C and D). Because t was not a single value, we interpolated values across the t-value range to generate ti (Dataset S1). We also assayed how fast ERK activity was lowered in response to U0126 addition and how fast it recovered after removal (Fig. S3 A and B). Because ERK inhibition occurred so rapidly after U0126 addition (15 min), ti values were not corrected for time required for drug response. We deduced the position at which reprogramming begins, Xoi, by using the equation Xoi = Xω − μti (Fig. 2E, blue bars; and Fig. S4). The Xoi values spanned germ cell rows 3 through 11 (Fig. 2E, blue bars; Dataset S1), with most mapping to rows 4 to 10 (Fig. 2E, blue bars). Therefore, germ-line sex reprogramming does not occur in the distal-most germ cells in the stem cell niche, but instead it begins after the germ cells have begun to move proximally to leave the niche.
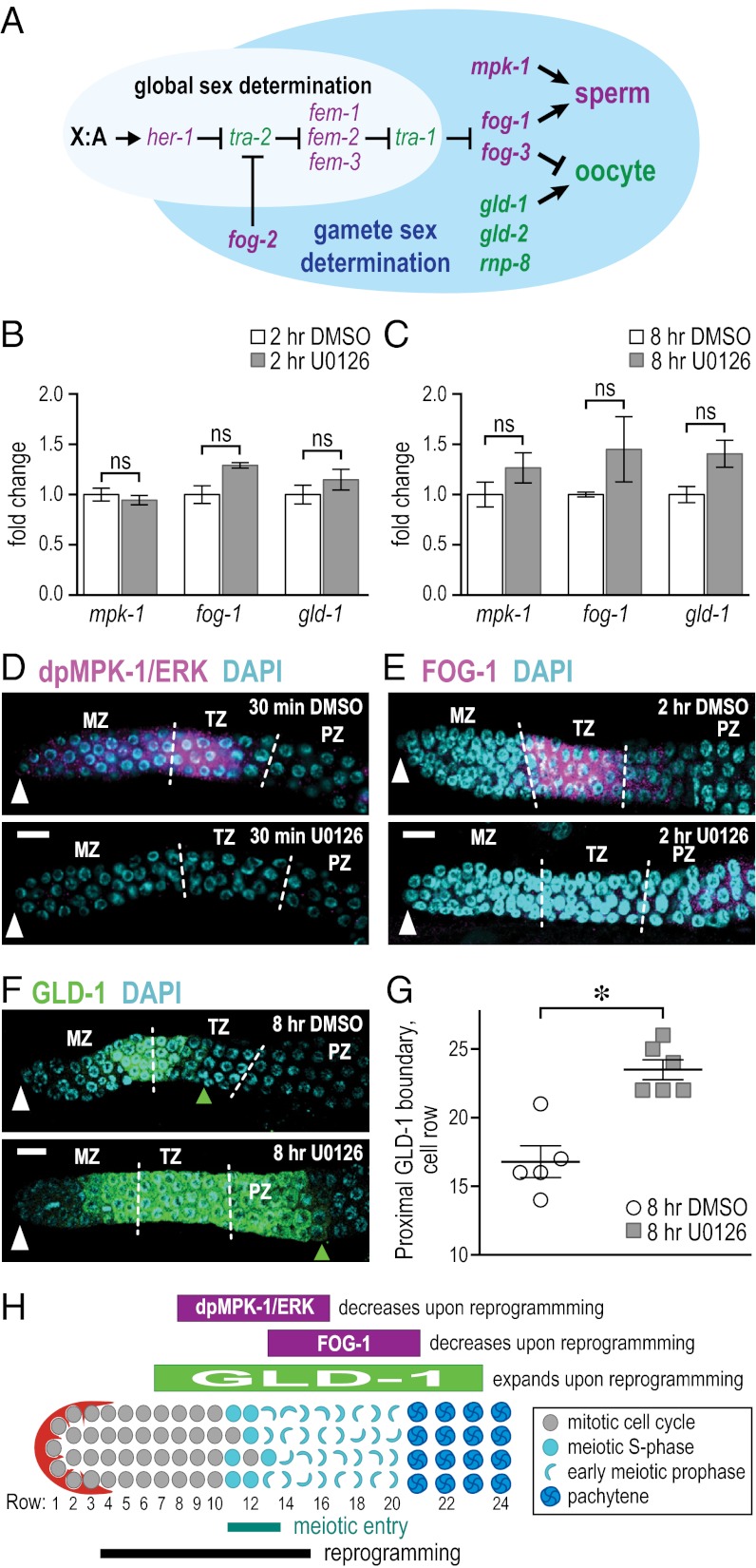
Fig. 3.
Chemical reprogramming alters key sperm–oocyte fate regulators. (A) Simplified pathway of gamete sex determination [adapted from Kimble and Crittenden (3)]. Sperm-promoting genes are shown in purple; oocyte-promoting genes are shown in green (5, 20). Arrows denote positive regulation; bars denote negative regulation. (B and C) Quantitative PCR of mRNAs encoding sperm/oocyte regulators after treatment with the DMSO solvent control (white bars) or the U0126 MEK inhibitor (gray bars) for 2 h (B) or 8 h (C). Each bar shows mean ± SEM, derived from three independent experiments; ns, no statistical significance (P > 0.05, Student t test). (D–F) Representative staining of activated MPK-1 (i.e., dpMPK-1), FOG-1, and GLD-1 proteins. For each regulator, the DMSO and U0126 patterns were typical of spermatogenic and oogenic germ lines, respectively (19, 22, 23). Conventions are as in Fig. 1. (Scale bars, 10 μm.) (D) Activated MPK-1/ERK protein was most abundant in the proximal MZ and distal TZ of DMSO-treated controls (Upper, n = 4). U0126 treatment dramatically reduced the abundance of activated MPK-1 (Lower, n = 4). (E) FOG-1 protein was abundant in proximal MZ, TZ, and distal PZ in DMSO-treated control animals (Upper, n = 3), but was undetectable after 2 h U0126 treatment (Lower, n = 6). (F) GLD-1 protein peaked at the MZ/TZ boundary of DMSO-treated controls (Upper). U0126 treatment expanded GLD-1 into pachytene zone (Lower). Green triangle marks proximal GLD-1 boundary. (G) Graph of the proximal GLD-1 boundary at 8 h DMSO treatment (n = 5) or 8 h U0126 treatment (n = 6). *P < 0.05, Student t test. (H) Summary schematic showing positions of reprogramming and meiotic entry along with positions of sperm–oocyte fate regulator changes.
Fig. 2.
Sites of sperm–oocyte fate reprogramming and meiotic entry. (A) Untreated (Left) and reprogrammed (Right) puf-8;lip-1 germ lines. To map the location of cells being reprogrammed, variables μ, t, and Xω were measured in U0126-treated germ lines. Conventions are as in Fig. 1. (B) Plot of proximal-most position of EdU incorporation, the EdU+ “front,” as a function of time after the 30 min pulse of EdU. The slope provides the rate of germ cell movement, μ. (C) Plot of percent animals with a mature oocyte (% oocyte-positive) vs. time to appearance of first oocyte. This plot was used to generate ti (Fig. S4). (D) Representative germ lines after 24 h DMSO (Left) or U0126 (Right) treatment. Germ lines were stained with DAPI (blue) for nuclei, SP56 (purple) for sperm, and OMA-2 (green) for oocytes. Green arrowhead indicates position of first oocyte, Xω. (Scale bar, 10 μm.) (E) Plot of percent reprogramming as a function of germ cell position. Blue bars, percent at each position that have initiated reprogramming; red bars, percent at each position that have completed reprogramming. Each bar shows mean ± SEM. The text and Figs. S3–S5 include mapping details. (F) Summary of data used to map the location of cells entering the meiotic cell cycle to rows 11 to 13. Reprogramming region is included for comparison with color coding as in E.
In addition to mapping where chemical reprogramming begins within the germ line, we also mapped where reprogramming finishes. To this end, we first determined Δ, the length of time ERK must be inhibited to accomplish reprogramming. Specifically, we measured the length of a U0126 pulse necessary for reprogramming and corrected that pulse length for the time required to recover ERK activity after drug removal (Fig. S5 A and B). Animals were treated with increasingly long U0126 pulses and then scored for sperm–oocyte fate reprogramming after 24 h from initial drug exposure to permit the generation of mature oocytes. An 8-h U0126 pulse reprogrammed all animals, whereas pulses as short as 2 h also worked, but in fewer animals (Fig. S5A). When corrected for the 1-h time of ERK recovery, we estimated that a Δ of 3 to 9 h ERK inhibition was required for reprogramming (Fig. S5B). Because Δ was not a single value, we interpolated values across the Δ-value range, or Δi (Dataset S2), and applied equation Xαi = Xω − μ(ti − Δi) to estimate Xαi, the position at which germ cells had completed reprogramming (Fig. 2E, red bars; and Fig. S5C). The Xαi values spanned rows 11 through 15, with most Xαi falling into rows 11 to 13 (Fig. 2E, red bars; and Dataset S2). A conservative estimate, gleaned from the extents of Xoi and Xαi, indicates that reprogramming of gamete sex is likely initiated between cell rows 3 and 11 and completed between rows 11 and 15 in the distal germ line (Fig. 2E). It is important to note that these values are rough estimates, as they cover reprogramming in all germ cells, not a single germ cell, and use variables (e.g., Xω, μ, ti, and Δ) from multiple experiments.
To relate the region of germ-line sex reprogramming to the site of meiotic entry, we next mapped where germ cells transition from the mitotic to the meiotic cell cycle in puf-8;lip-1 mutants. That transition is best gleaned by using the combined results of a series of markers. Staining with the antibody to phosphohistone H3, a marker of mitotic M-phase, was restricted to rows 1 to 12, with the vast majority in rows 1 to 10 (Fig. 2F and Fig. S6 A and B). Labeling with a 30-min pulse of thymidine analogue EdU, a marker incorporated into DNA during mitotic and meiotic S-phase, was limited to rows 1 to 14 (Fig. 2F and Fig. S6C). Chromosomal HIM-3 staining (HIM-3 is a marker of meiotic entry) extended proximally from row 11 (Fig. 2F and Fig. S6D). Cells in early meiotic prophase, detected as crescent-shaped nuclei after DAPI staining, spanned rows 13 to 20 (Fig. 2F and Fig. S6A). These findings together indicate that most puf-8;lip-1 germ cells are maintained in the mitotic cell cycle until row ∼10, that most enter meiotic S-phase at row ∼11, and that most enter meiotic prophase at row ∼13. By comparison, our estimate of the sperm–oocyte reprogramming region spanned rows 3 to 15. Therefore, gamete sex reprogramming appears to begin in mitotically dividing germ cells and to be completed in early meiotic prophase germ cells. The coincidence of meiotic entry with the sperm–oocyte fate decision was inferred previously (6), as was the distal germ line as the site of the sperm–oocyte decision (10, 14, 15). Our results are consistent with those inferences and support the notion that chemical reprogramming of the sperm–oocyte fate decision is likely to reflect the normal sperm–oocyte fate decision.
We conclude that chemical reprogramming of the sperm–oocyte decision occurs in a region spanning the proximal MZ and distal transition zone (TZ), and suggest that commitment to sperm or oocyte development normally occurs in the same region. Consistent with this physical mapping, previous studies found that germ cells in the proximal MZ are actively transitioning from a stem cell-like state to a differentiated state (16). Intriguingly, germ cells in the MZ are also uniquely susceptible to reprogramming into neurons after removal of just one chromatin factor (17). Perhaps germ cells in the proximal MZ have acquired a “transient regulatory state” (18), which is particularly vulnerable to reprogramming.
Key Sperm–Oocyte Fate Regulators Change Abundance upon Chemical Reprogramming.
The sperm-to-oocyte fate reprogramming of puf-8;lip-1 mutants relies on established germ-line sex regulators, including ERK/MPK-1 and FOG-1 (4, 6, 9). To investigate the effect of reprogramming on the sex-specific expression of these and other germ-line sex regulators, we assayed their transcript and protein abundance, focusing on regulators at the end of the germ-line sex determination pathway (Fig. 3A). By quantitative PCR, the mRNA abundance of all eight regulators assayed (mpk-1, fog-1, gld-1, fog-3, tra-1, fem-3, gld-2, and rnp-8) was essentially unchanged at 2 and 8 h after U0126 addition compared with control (Fig. 3 B and C and Fig. S7 B and C). Therefore, reprogramming occurs without a significant change in abundance of transcripts encoding these key terminal sperm–oocyte fate regulators.
We did, however, find abundance changes in three sperm–oocyte fate regulatory proteins: the sperm-promoting form of MPK-1/ERK [diphosphorylated MPK-1 (dpMPK-1)] (19), sperm-promoting FOG-1 (6), and oocyte-promoting GLD-1 (20). Activated dpMPK-1 was easily detectable in the TZ of spermatogenic DMSO-treated control germ lines (Fig. 3D, Upper), a pattern typical of spermatogenic germ lines (19), but was barely detectable in U0126-treated germ lines 30 min after drug treatment (Fig. 3D, Lower), a pattern typical of oogenic germ lines (19). This U0126 effect on dpMPK-1 was predicted: U0126 inhibits MEK (21) and activates MPK-1 to its diphosphorylated form. The decrease in dpMPK-1 upon drug addition is consistent with our previous finding that U0126 induces chemical reprogramming specifically through inhibition of Ras-ERK signaling in puf-8;lip-1 mutants (4).
In addition, FOG-1 protein decreased sharply in the TZ within 2 h of U0126 addition (Fig. 3E), changing from a typical spermatogenic pattern to a typical oogenic pattern of FOG-1 expression (22). Finally, the oocyte-promoting GLD-1 protein expanded dramatically within 8 h of treatment from a small patch at the MZ/TZ border in DMSO-treated germ lines to a large area extending into the pachytene zone of U0126-treated germ lines (Fig. 3 F and G). This change again shifts from a spermatogenic to an oogenic pattern of GLD-1 expression (23). The U0126-induced changes in FOG-1 and GLD-1 likely reflect the downstream readout of sperm-to-oocyte reprogramming. We conclude that chemical reprogramming induces dramatic changes in three major regulators of germ-line sex determination.
The striking changes in sperm–oocyte fate regulators observed upon chemical reprogramming strongly support the idea that reprogramming operates through the normal sperm–oocyte cell fate machinery, as previously suggested (4). The mechanism inducing these changes is likely exerted at a posttranscriptional or posttranslational level, because corresponding mRNAs did not change in abundance. We speculate that MPK-1/ERK activity may exert its effect on the sperm–oocyte decision by controlling terminal germ-line sex regulators. We also note that reprogramming-induced changes of germ-line sex regulators were most impressive in germ cells that had entered early meiotic prophase, consistent with our findings that the sperm–oocyte decision can be reprogrammed after entry into the meiotic cell cycle and that the physical map of reprogramming extends into the meiotic zone.
Conclusion
A longstanding question in metazoan germ cell biology has been whether the mitosis–meiosis and sperm–oocyte cell fate choices are one or two regulatory events (see the Introduction). We have found that the mitosis–meiosis and sperm–oocyte decisions are separable and therefore cannot be a single event. This conclusion seems intuitive for organisms whose regulators of meiotic entry act in both males and females (24–26). However, this conclusion has been difficult to confirm experimentally because the regulatory pathways underlying the two decisions are intimately coupled, with meiotic entry regulators affecting the sperm–oocyte decision and vice versa. For example, FOG-1 and FOG-3 are not only major regulators of sperm fate specification, but they also have a role in the mitosis–meiosis decision (6, 15, 27, 28). Similarly, GLD-1 and GLD-2 are not only major regulators of meiotic entry, but they also promote the oocyte fate (24, 29). A closely linked relationship also holds true for mitosis–meiosis and mating type regulation in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe (e.g., refs. 30–34). Given the theme of coupled but separate regulation in C. elegans and the yeasts, we propose that the mitosis–meiosis and sperm–oocyte cell fate choices are likely to be two regulatory events in all germ cells.
Materials and Methods
Strains.
Nematodes were maintained as described previously (35). puf-8(q725);lip-1(zh15) homozygotes were picked from a strain containing balancer mIn1[mIs14 dpy-10(e128)]. glp-1(q224 ts);puf-8(q725);lip-1(zh15) animals were obtained similarly.
Chemical Reprogramming.
Chemical treatments were performed as described previously (4). Briefly, nematodes were treated in liquid culture, including S-Media and OP50 Escherichia coli, as indicated. Differential interference contrast microscopy was used to monitor the percentage of germ lines with oocytes after treatments.
Germ Cell Position and Germ-Line Regions.
Position within the germ line was measured in gcds from the distal end as described previously (36).
Temperature Shifts.
glp-1(q224ts);puf-8(q725);lip-1(zh15) animals were collected at L1 arrest and grown to the mid-L4 larval stage at 15 °C. To initiate the temperature shift, animals were picked into 25 °C M9 buffer for ∼5 min, and transferred to 25 °C nematode growth media plates for the indicated times. Chemical reprogramming of glp-1(q224ts);puf-8(q725);lip-1(zh15) animals was conducted as described earlier, except animals were maintained at 25 °C.
Immunohistochemistry.
Germ lines were extruded in M9/0.2 mM levamisole/0.1% Tween 20 and fixed in 3% (vol/vol) paraformaldehyde/PBS solution/0.1% Tween 20 for 15 min at room temperature, followed by a 15-min postfix in −20 °C methanol. Germ lines were equilibrated in PBS solution/0.1% Tween 20 for 15 min at room temperature and blocked by 1 h incubation in 0.5% BSA/PBS solution/0.1% Tween 20. Primary antibodies were incubated overnight at 4 °C. Primary antibodies were diluted in 0.5% BSA/PBS solution/0.1% Tween 20 at dilutions of: 1:200 mouse monoclonal anti-pTEpY ERK-1 (Sigma-Aldrich), 1:100 rabbit anti–HIM-3 (13), 1:100 mouse anti-phosphohistone H3 (Cell Signaling), 1:4 rat anti–FOG-1 N terminus (15), 1:50 rabbit anti–OMA-2 (37), 1:100 rabbit anti–GLD-1 (38), and 1:50 mouse anti-SP56 (39). Secondary antibodies (Cy3 anti-rabbit, Cy3 anti-rat, Cy3 anti-mouse, Cy5 anti-mouse) were obtained from Jackson ImmunoResearch and were used at 1:500 dilutions in 5% (vol/vol) BSA/PBS solution/0.1% Tween 20. Germ cell nuclei were visualized by DAPI staining. Fluorescence intensities were measured with the program ImageJ (40) . For comparisons of immunohistochemistry fluorescence, controls and samples were prepared in parallel and imaged in the same microscope session with all microscope settings kept identical.
Western Blot Analysis.
Semiquantitative Western blotting was performed as described previously (4). Briefly, active diphosphorylated MPK-1/ERK was detected with 1:5,000 mouse monoclonal anti-pTEpY ERK-1 (Sigma-Aldrich) and 1:10,000 HRP-conjugated anti-mouse (Jackson ImmunoResearch). After stripping with Restore Plus Western Blot Stripping buffer (Pierce), blots were reblocked and total MPK-1/ERK was detected with 1:20,000 rabbit anti–ERK-1/2 (Sc94; Santa Cruz Biotechnology) and 1:10,000 HRP-conjugated anti-rabbit (Jackson ImmunoResearch). Blots were restripped and reblocked, and ∝-tubulin was detected with 1:10,000 mouse monoclonal anti–∝-tubulin (Sigma-Aldrich) and 1:10,000 HRP-conjugated anti-mouse. The relative activity of MPK-1/ERK was determined by normalizing active MPK-1/ERK signal and total MPK-1/ERK signal to ∝-tubulin with the program ImageJ (40).
EdU Labeling.
The Click-iT EdU Imaging Kit (Invitrogen) was used to label DNA via incorporation of a “clickable” thymidine analogue, EdU (12). For germ cell rate of movement experiments, labeling was performed by feeding animals EdU-labeled E. coli for 30 min, followed by growth in liquid culture containing 50 μM U0126 in DMSO (or DMSO alone), S-media, and OP50 E. coli. For glp-1(q224 ts);puf-8(q725);lip-1(zh15) experiments, animals were labeled by incubation for 30 min in M9 buffer containing 100 μM EdU. Germ lines were then extruded and fixed, and the incorporated EdU was visualized after conjugation to Alexa Fluor 488 azide. Total DNA was visualized by DAPI staining.
Quantitative PCR.
Total RNA was isolated from worm extract using TRIzol (Invitrogen) and the RNeasy Micro Kit (Qiagen) following the manufacturer’s protocols. Oligo-dT–primed cDNA was prepared from total RNA samples by using a SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) following manufacturer’s protocols. To quantify specific transcript levels, quantitative PCR was performed by using TaqMan Gene Expression Master Mix (Applied Biosystems) and TaqMan Gene Expression Assays (Applied Biosystems) in a 7500 Fast Real-Time PCR System (Applied Biosystems). All assays were normalized to the endogenous control, eft-3. The following TaqMan assays were used: fog-1(L), Ce02415381_g1; fog-3, Ce02412829_g1; gld-1, Ce02409901_g1; tra-1, Ce02407051_g1; fem-1, Ce02463926_g1; fem-3, Ce02457444_g1; gld-2, Ce02408169_g1; rnp-8, Ce02413620_g1; eft-3, Ce02448437_gH; and mpk-1, Ce02445290_m1.
Supplementary Material
Acknowledgments
We thank Anne Helsley and Laura Vanderploeg for help in manuscript and figure preparation; Dr. R. Lin (University of Texas Southwestern Medical Center) and Dr. S. Ward (University of Arizona) for OMA-2 and SP56 antibodies, respectively; Elena Sorokin for reading the manuscript; and all members of the J.K. laboratory for many helpful discussions. This work was supported by National Institutes of Health Grant GM069454 (to J.K.). J.K. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300928110/-/DCSupplemental.
References
- 1.Kimble J, Page DC. The mysteries of sexual identity. The germ cell’s perspective. Science. 2007;316(5823):400–401. doi: 10.1126/science.1142109. [DOI] [PubMed] [Google Scholar]
- 2.Kimble J. Molecular regulation of the mitosis/meiosis decision in multicellular organisms. Cold Spring Harb Perspect Biol. 2011;3(8):a002683. doi: 10.1101/cshperspect.a002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- 4.Morgan CT, Lee MH, Kimble J. Chemical reprogramming of Caenorhabditis elegans germ cell fate. Nat Chem Biol. 2010;6(2):102–104. doi: 10.1038/nchembio.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis R, Schedl T. Sex determination in the germ line (March 5, 2007) 2007. WormBook, ed The C. elegans Research Community, WormBook, 10.1895/wormbook.1.82.2. Available at www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 6.Barton MK, Kimble J. fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics. 1990;125(1):29–39. doi: 10.1093/genetics/125.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P-J, Singal A, Kimble J, Ellis RE. A novel member of the tob family of proteins controls sexual fate in Caenorhabditis elegans germ cells. Dev Biol. 2000;217(1):77–90. doi: 10.1006/dbio.1999.9521. [DOI] [PubMed] [Google Scholar]
- 8.Barton MK, Schedl TB, Kimble J. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics. 1987;115(1):107–119. doi: 10.1093/genetics/115.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee M-H, et al. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics. 2007;177(4):2039–2062. doi: 10.1534/genetics.107.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachorik JL, Kimble J. Redundant control of the Caenorhabditis elegans sperm/oocyte switch by PUF-8 and FBF-1, two distinct PUF RNA-binding proteins. Proc Natl Acad Sci USA. 2005;102(31):10893–10897. doi: 10.1073/pnas.0504593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berset T, Hoier EF, Battu G, Canevascini S, Hajnal A. Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science. 2001;291(5506):1055–1058. doi: 10.1126/science.1055642. [DOI] [PubMed] [Google Scholar]
- 12.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105(7):2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zetka MC, Kawasaki I, Strome S, Müller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999;13(17):2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clifford R, et al. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development. 2000;127(24):5265–5276. doi: 10.1242/dev.127.24.5265. [DOI] [PubMed] [Google Scholar]
- 15.Thompson BE, et al. Dose-dependent control of proliferation and sperm specification by FOG-1/CPEB. Development. 2005;132(15):3471–3481. doi: 10.1242/dev.01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cinquin O, Crittenden SL, Morgan DE, Kimble J. Progression from a stem cell-like state to early differentiation in the C. elegans germ line. Proc Natl Acad Sci USA. 2010;107(5):2048–2053. doi: 10.1073/pnas.0912704107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331(6015):304–308. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertrand V, Hobert O. Lineage programming: navigating through transient regulatory states via binary decisions. Curr Opin Genet Dev. 2010;20(4):362–368. doi: 10.1016/j.gde.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee M-H, et al. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007;3(12):e233. doi: 10.1371/journal.pgen.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim KW, et al. Antagonism between GLD-2 binding partners controls gamete sex. Dev Cell. 2009;16(5):723–733. doi: 10.1016/j.devcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncia JV, et al. MEK inhibitors: the chemistry and biological activity of U0126, its analogs, and cyclization products. Bioorg Med Chem Lett. 1998;8(20):2839–2844. doi: 10.1016/s0960-894x(98)00522-8. [DOI] [PubMed] [Google Scholar]
- 22.Lamont LB, Kimble J. Developmental expression of FOG-1/CPEB protein and its control in the Caenorhabditis elegans hermaphrodite germ line. Dev Dyn. 2007;236(3):871–879. doi: 10.1002/dvdy.21081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones AR, Francis R, Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol. 1996;180(1):165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- 24.Kadyk LC, Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125(10):1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- 25.Menke DB, Koubova J, Page DC. Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev Biol. 2003;262(2):303–312. doi: 10.1016/s0012-1606(03)00391-9. [DOI] [PubMed] [Google Scholar]
- 26.Anderson EL, et al. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci USA. 2008;105(39):14976–14980. doi: 10.1073/pnas.0807297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis RE, Kimble J. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics. 1995;139(2):561–577. doi: 10.1093/genetics/139.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snow JJ, Lee MH, Verheyden J, Kroll-Conner PL, Kimble J. C. elegans FOG-3/Tob can either promote or inhibit germline proliferation, depending on gene dosage and genetic context. Oncogene. 2012 doi: 10.1038/onc.2012.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim KW, Wilson TL, Kimble J. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc Natl Acad Sci USA. 2010;107(40):17445–17450. doi: 10.1073/pnas.1012611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harigaya Y, Yamamoto M. Molecular mechanisms underlying the mitosis-meiosis decision. Chromosome Res. 2007;15(5):523–537. doi: 10.1007/s10577-007-1151-0. [DOI] [PubMed] [Google Scholar]
- 31.Kassir Y, et al. Transcriptional regulation of meiosis in budding yeast. Int Rev Cytol. 2003;224:111–171. doi: 10.1016/s0074-7696(05)24004-4. [DOI] [PubMed] [Google Scholar]
- 32.Kassir Y, Simchen G. Regulation of mating and meiosis in yeast by the mating-type region. Genetics. 1976;82(2):187–206. doi: 10.1093/genetics/82.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell AP. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58(1):56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto M. The molecular control mechanisms of meiosis in fission yeast. Trends Biochem Sci. 1996;21(1):18–22. [PubMed] [Google Scholar]
- 35.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan DE, Crittenden SL, Kimble J. The C. elegans adult male germline: Stem cells and sexual dimorphism. Dev Biol. 2010;346(2):204–214. doi: 10.1016/j.ydbio.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Detwiler MR, Reuben M, Li X, Rogers E, Lin R. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev Cell. 2001;1(2):187–199. doi: 10.1016/s1534-5807(01)00026-0. [DOI] [PubMed] [Google Scholar]
- 38.Jan E, Motzny CK, Graves LE, Goodwin EB. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 1999;18(1):258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward S, Roberts TM, Strome S, Pavalko FM, Hogan E. Monoclonal antibodies that recognize a polypeptide antigenic determinant shared by multiple Caenorhabditis elegans sperm-specific proteins. J Cell Biol. 1986;102(5):1778–1786. doi: 10.1083/jcb.102.5.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasband WS. 2009. ImageJ (National Institutes of Health, Bethesda, MD). Available at http://rsb.info.nih.gov/ij/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.