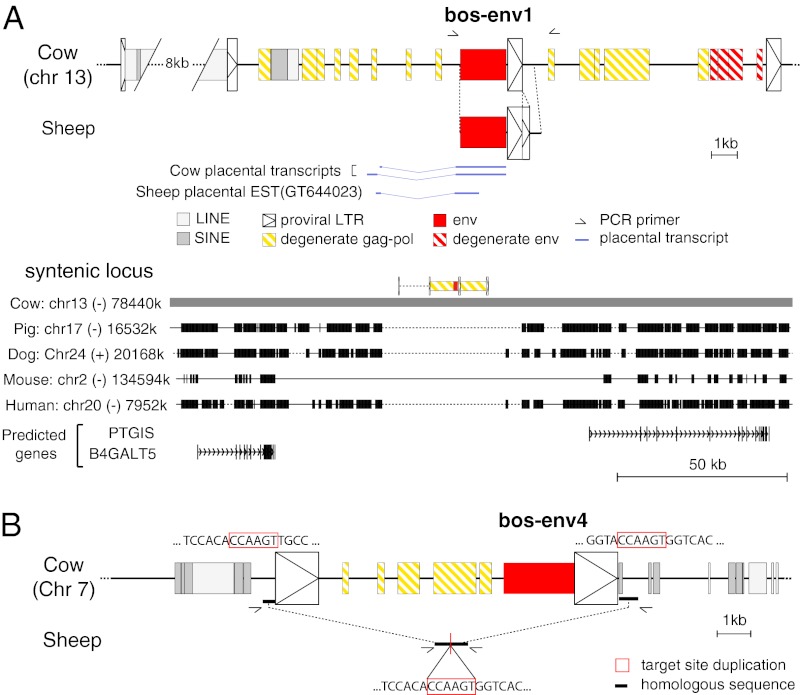
Fig. 5.
Characterization of the bos-env1–, ovis-env1–, and bos-env4–containing ERVs and of their genomic location in the cow and sheep. (A) Characterization of the bos-env1–containing ERV. (Upper) Structure of the bos-env1–containing ERV and evidence for orthology between the cow and sheep env1 sequences. Homologous regions common to both sequences are aligned. Repeated mobile elements (gray) as identified by the RepeatMasker web program are positioned. Of note, bos-env1 is in a provirus directly followed by a tandem repeat of a homologous provirus sharing a common LTR. The proviral LTRs, the degenerate gag-pol and env genes, and the env1 gene ORF sequence are indicated (the key for symbols used is shown below the panel). PCR primers used to identify the bos-env1 orthologous copy in the sheep (black half arrows) and splice sites for the env subgenomic transcripts as determined by RT-PCR of cow placental RNA or by alignment with an EST sequence for the sheep are indicated. The nonhomologous sequence in the sheep LTR positioned 3′ to the env1 sequence compared with the cow corresponds to a 200-bp direct tandem duplication of part of the LTR 3′ end in the sheep. (Lower) Absence of the bos-env1–containing ERV in the genomes of distant mammalian lineages. The bos-env1–containing provirus (in yellow, with its LTRs schematized by boxed triangles and the env gene in red) was used as a reference, and synteny between the cow, pig, dog, mouse, and human genomes was determined with the Comparative Genomic tool of the UCSC Genome Browser (http://genome.ucsc.edu/). The positions of exons (vertical lines) of the resident B4GALT5 and PTGIS gene and the sense of transcription (arrows) are indicated. Homologous regions are shown as black boxes, nonhomologous regions as thin lines (not to scale), and gaps as dotted lines. (B) Structure of the bos-env4–containing ERV and evidence of the absence of proviral integration in the sheep syntenic locus. The symbol code is as in A. Homologous sequences flanking the provirus in the cow and colinear in the sheep are indicated by bold lines. Comparison of the flanking sequences of the cow provirus with those of the empty locus in the sheep (obtained by PCR with the indicated primers) provides evidence for target-site duplication in the cow (red boxes), a characteristic feature of retroviral integration.