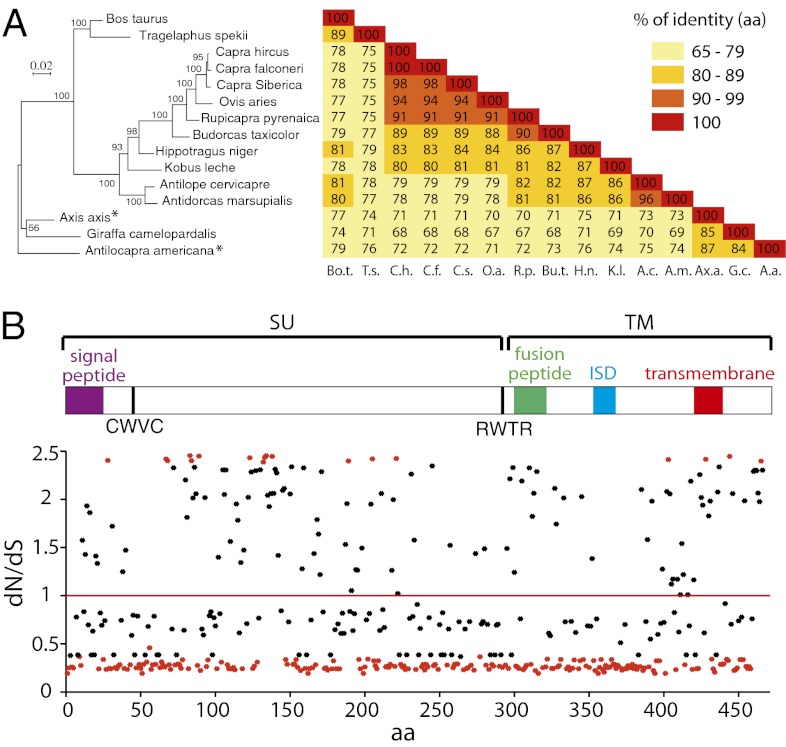
Fig. 10.
Evidence for sequence conservation of and selection pressure on the syncytin-Rum1 gene. (A, Left) Syncytin-Rum1–based maximum-likelihood phylogenetic tree determined using amino acid alignment of the Syncytin-Rum1 proteins identified in Fig. 9, inferred with the RAxML program. The horizontal branch length and scale indicate the percentage of amino acid substitutions. Percent bootstrap values obtained from 1,000 replicates are indicated at the nodes. An asterisk indicates that the sequence used for Axis axis is a consensus of the four full-length gene ORFs (otherwise branching together) and, for Antilocapra americana, the one shown to be fusogenic in the pseudotyping assay (Fig. 9). (A, Right) Double-entry table for the pairwise percentage of amino acid sequence identity between the syncytin-Rum1 genes belonging to the indicated species. (B) Site-specific analysis of selection on syncytin-Rum1 gene codons, using the PAML (M8 model) package. The relevant selection indexes are provided for each codon. A schematic representation of the Syncytin-Rum1 protein domains is given also; conventions are as in Fig. 2. Significant values (P ≥ 0.95) are represented as red dots.