Figure 3.
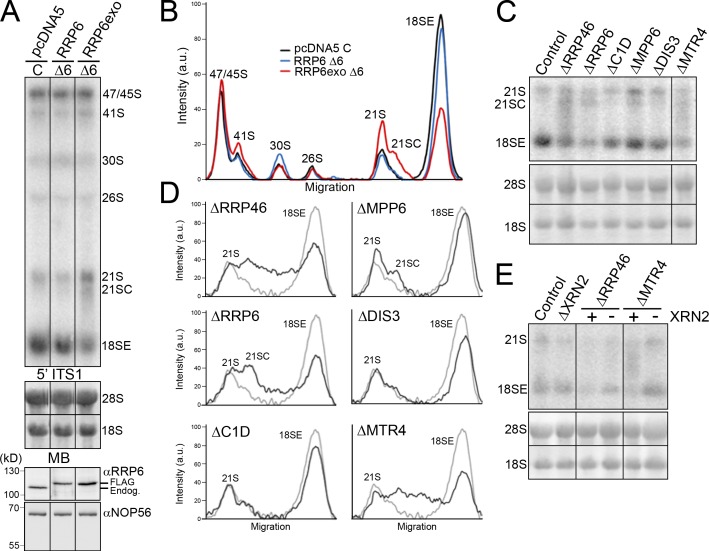
The exonuclease activity of RRP6, the exosome core, and the RNA helicase MTR4 are required for 18SE production. (A) HEK293 cells stably transfected with plasmids expressing the FLAG tag alone (pcDNA5), FLAG-RRP6, or FLAG-RRP6exo were transfected with either control siRNAs (C) or siRNAs that target the endogenous RRP6 mRNA (Δ6). The FLAG-RRP6 cDNA sequences had been altered to render the mRNA resistant to the RRP6 siRNAs. Levels of the endogenous and plasmid-expressed RRP6 were monitored by Western blotting (bottom panels). Levels of pre-rRNAs were analyzed by Northern blotting using a probe hybridizing to the 5′ end of ITS1. (B) The levels of the pre-rRNA intermediates from A, which is representative of two experiments, are plotted. The identity of each peak is indicated. (C) RNA extracted from HeLa cells depleted of exosome or exosome-cofactor proteins (as indicated above each lane) by RNAi was analyzed by Northern blotting using a probe specific for the 5′ end of ITS1. (D) The levels of the pre-rRNA intermediates from C, which is representative of more than three experiments, are plotted for each knockdown (black) and the control (gray). The identity of each peak is indicated. (E) RNA extracted from control HeLa cells or HeLa cells depleted of RRP46 or MTR4, alone or together with XRN2 (indicated at top), was analyzed by Northern blotting using a probe against the 5′ end of ITS1. Positions of the mature and pre-rRNAs are indicated on the left. RNA loading was monitored by methylene blue staining of the 18S and 28S rRNAs (MB). In codepletion experiments, the presence (+) or depletion (−) of XRN2 is indicated. Black lines in A, C, and E indicate intervening lanes that have been removed.